Antibody structure function and diversity Lymphocyte Maturation Overview

Antibody structure, function and diversity

Lymphocyte Maturation
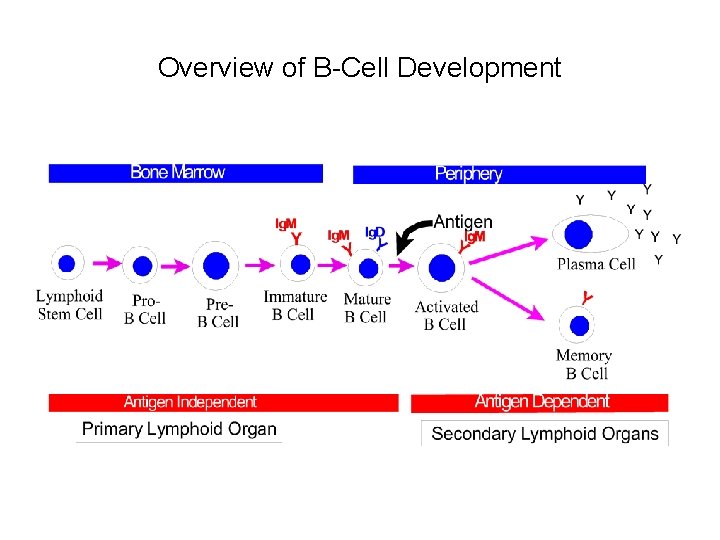
Overview of B-Cell Development
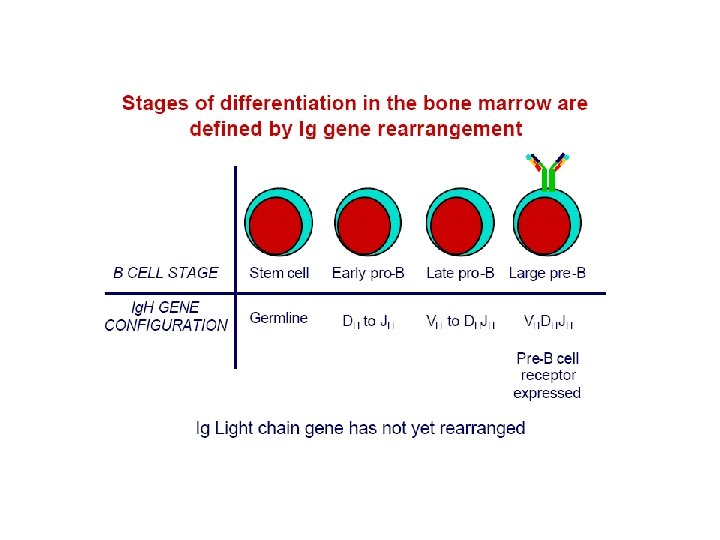
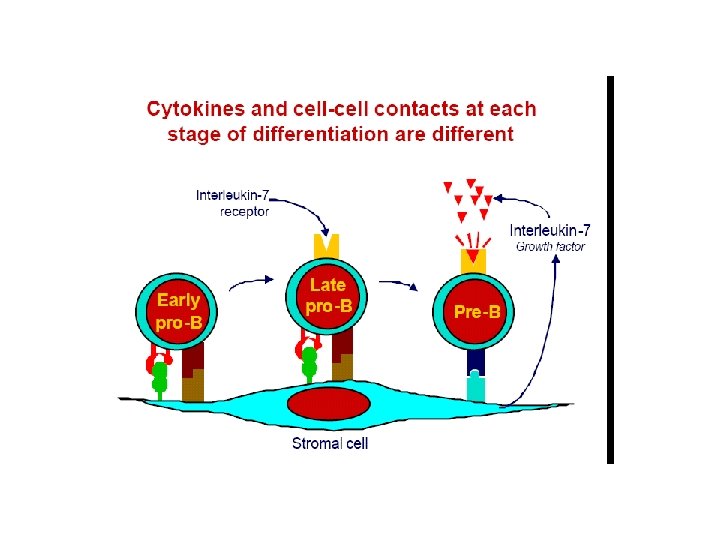

Lymph. maturation • Maturation of lymphocytes from bone-marrow stem cells consist of 3 types of processes: • proliferation of immature cells • expression of ag receptor genes • selection of lymphocytes that express useful ag receptors. • B cell maturation depends on the rearrangement of the Ig DNA in the haemapoetic stem cells
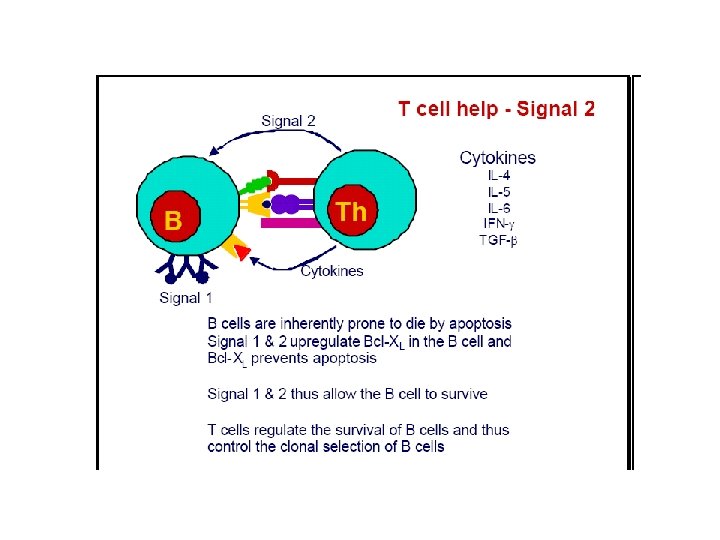
B cell activation • Depending on nature of ag , proceeds by two different routes: • 1 -Dependent upon Th cells(TD) • 2 -Independent of Th cells (TI) • B cell response to TD ags requires direct contact with Th cells, not simply exposure to Th derived cytokines.
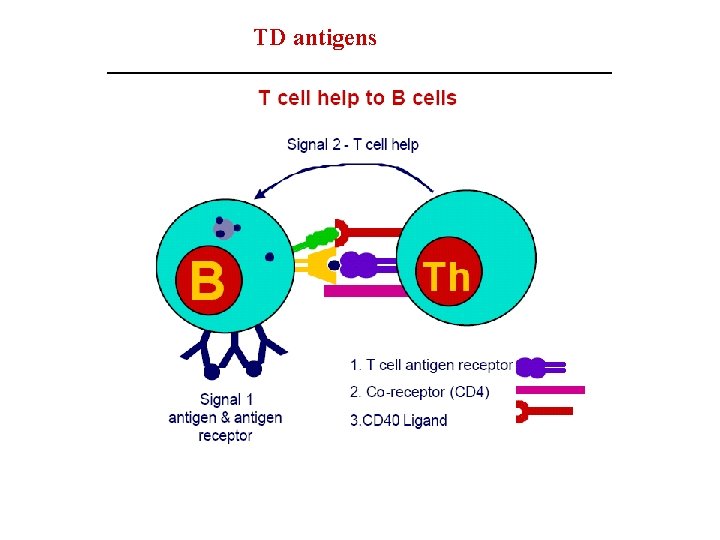
TD antigens
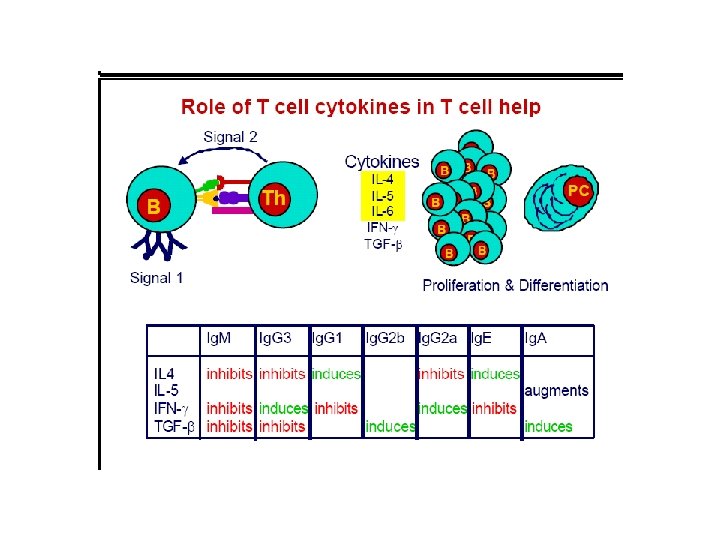
T independent antigen • TI ags divided into TI-1 & TI-2 • TI-2 are highly repetitious molecules such as bacterial cell wall polysaccharides with repeating polysacc. Units, activate B cells by extensively cross-linking s. Ig receptors. • Most TI -1 ags are polyclonal activators(mitogens) that activate Bcells regardless of antigenic specificity. LPS at low conc. Stimulates specific abs. High conc. Is a polyclonal activator.
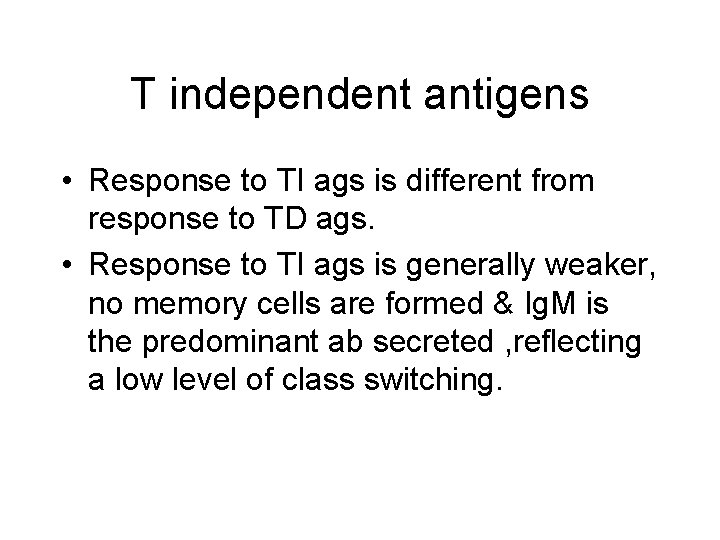
T independent antigens • Response to TI ags is different from response to TD ags. • Response to TI ags is generally weaker, no memory cells are formed & Ig. M is the predominant ab secreted , reflecting a low level of class switching.
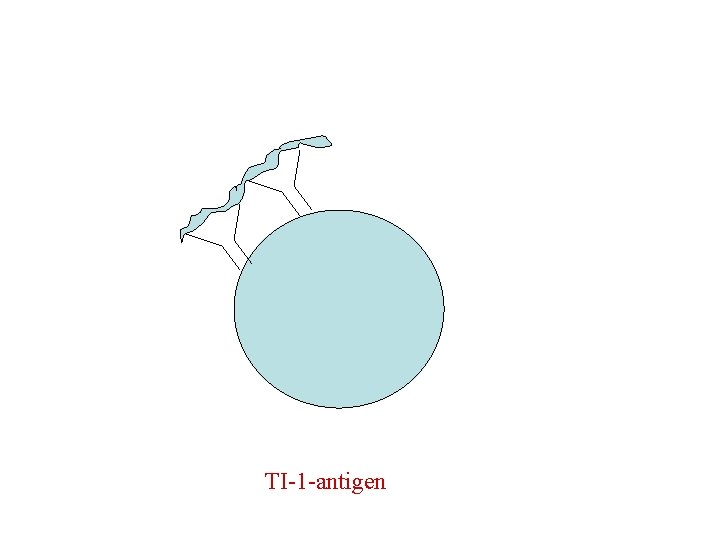
TI-1 -antigen
Humoral Immunity • Humoral immunity is mediated by antigenspecific proteins called antibodies. • The humoral immune response is uniquely adapted for elimination of extracellular pathogens. • Cells of the B-lineage are the principal cell types involved in humoral immunity.
Where else are antibodies found in the body? • In addition to the fluid part of blood, anti-bodies are found in several anatomic locations. – – Cytoplasm and cell surface of B lymphocytes Interstitial spaces Secretory fluids (mucous, tears, milk) Surface of certain immune effector cells which do not synthesize antibody but have specific receptors (Fc receptors that bind antibody molecules.
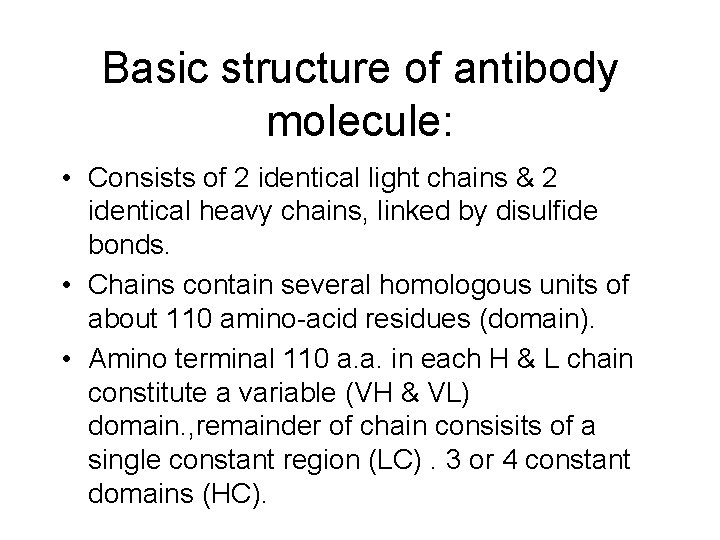
Basic structure of antibody molecule: • Consists of 2 identical light chains & 2 identical heavy chains, linked by disulfide bonds. • Chains contain several homologous units of about 110 amino-acid residues (domain). • Amino terminal 110 a. a. in each H & L chain constitute a variable (VH & VL) domain. , remainder of chain consisits of a single constant region (LC). 3 or 4 constant domains (HC).
STRUCTURAL BASIS OF ANTIBODY DIVERSITY • Antibodies are the antigen-specific products of B cells, and the production of antibody in response to infection is the main contribution of B cells to adaptive immunity. • The antibody molecule has two separate functions: one is to bind specifically to molecules from the pathogen that elicited the immune response; the other is to interact with various cells and molecules to destroy the pathogen once the antibody is bound to it. • These functions are structurally separated in the antibody molecule, one part of which specifically recognizes antigen and the other engages effector mechanisms that dispose of it. • The antigen-binding region varies extensively between antibody molecules and thus is known as the variable region or V region. • The region of the antibody molecule that engages the effector functions of the immune system does not vary in the same way and is thus called the constant region or C region.
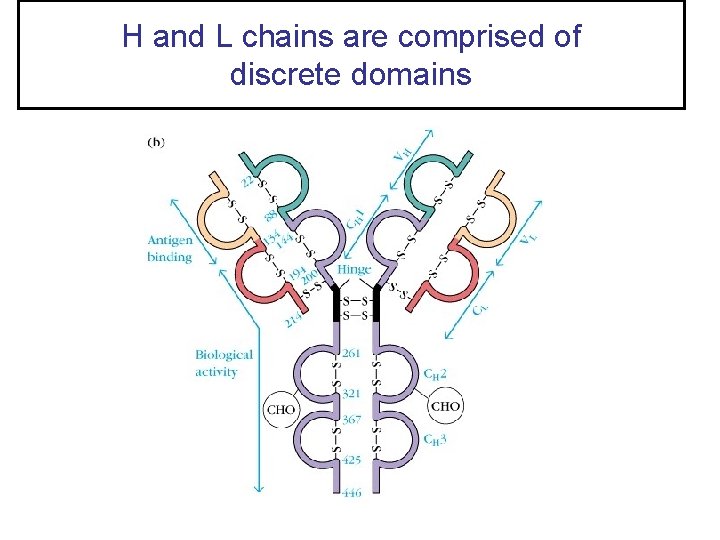
H and L chains are comprised of discrete domains
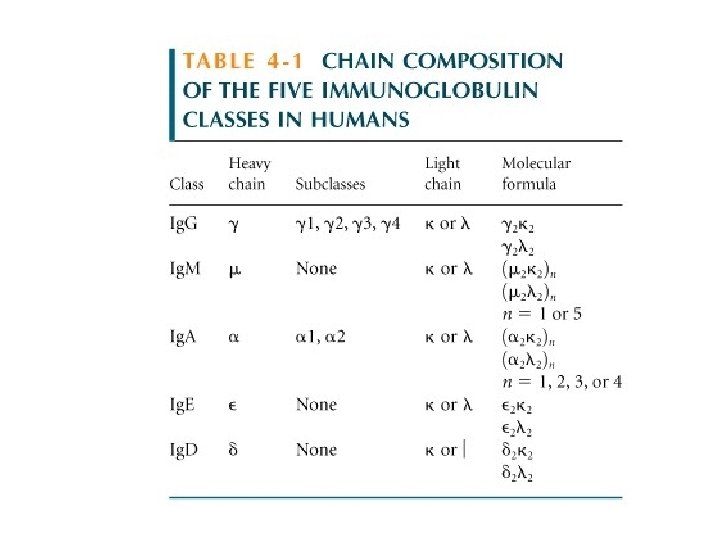
Immunoglobulin Sequencing Studies • Early sequencing studies done on myeloma proteins: the products of malignant Ab producing cells. • Light chain sequencing: – The amino terminal half of the L chains varied considerably between different isolates – termed the variable (V) region – The carboxyl terminal half of the L chain – called the constant (C) region – had two basic amino acid sequences – i. e. two types of light chains – kappa (κ) and lambda (λ). Subsequently it was shown that there are three – four subtypes of lambda chains – λ 1, λ 2, etc. • IMPORTANT NOTE: A single antibody molecule contains only one light chain type – never both.
Immunoglobulin Sequencing Studies • Heavy chain sequencing: – The amino terminal 110 amino acids make up the variable region – The remaining part of the H chains showed five basic sequence patterns corresponding to five different heavy-chain contstant (C) regions (γ, α, µ, δ, ε) – Each of these five different heavy-chains is called and isotype. – The constant regions of µ and ε are larger than the constant regions of γ, α, and δ due to the presence of an extra constant domain. • A single antibody molecule contains a single type of heavy-chain.
Immunoglobulin Domains • Both H and L chains contain several homologous units of about 110 amino acids termed a – DOMAIN. • Each domain contains an intrachain disulfide bond which forms a loop of approximately 60 amino acids. • L chains contain one variable domain (VL), and one constant domain (CL) • H chains contain one variable domain – (VH) – and either three or four constant domains – (CH 1, CH 2, CH 3, and CH 4) depending on the antibody class.
Variable-Region Domains • The amino-terminal domain of both H and L chains is termed a variable (V) region due to the discovery of extensive seequence divergence between different antibody proteins in this part of the molecule. These are designated VH and VL for H and L chains, respectively. • V regions have been demonstrated to be responsible for antigen specificity of the immunoglobulin. • Sequence analysis of many VH and VL domains revealed that variability is concentrated in several hypervariable (HV) regions. • There are three hypervariable regions in each VH and VL domain. • The stretches of amino acid between the HVs are termed framework regions and these exhibit far less variability. • The interaction between the HVs of the H and L chains form the antigen binding site of the antibody molecule. • Since these HVs fold into a structure that is complementary to the antigen epitope, the HVs are also called complementaritydetermining regions (CDRs).
Constant-Region Domains • The carboxyl-terminal domains of immunoglobulins display considerably less sequence variability within a given isotype than observed for V-region domains. These domains are referred to as constant (C) regions. • H-chain C regions are numbered (CH 1, CH 2, CH 3, and CH 4) beginning with the most V-region proximal domain. • The C region domains of the H-chain have been shown to be responsible for many aspects of antibody function, including interaction with Fc receptors, complement fixation, transplacental transfer, the ability to form multimers, and the capacity to be secreted across mucosal surfaces. • Because different H-chain isotypes have different C region domains, these capabilities vary with the class of the particular antibody. • Five major classes of H-chain C regions exist: (γ, α, µ, δ, ε). As a direct consequence of the correlation between the H-chain class of an antibody and its resultant effector functions, immunoglobulins have been named according to there heavy chain, i. e. , Ig. G, Ig. A, Ig. M, Ig. D and Ig. E.
Hinge Region • Immunoglobulins have a hinge region located C-terminal of the CH 1 domain of their H-chains. • In the case of the H-chains of the µ and ε isotypes the hinge is so elongated that it is actually an extra immunoglobulin domain explaining the presence of a fourth C domain in these isotypes. • Other heavy chain isotypes (γ, α, and δ) use shorter stretches of polypeptide, which are thought nonetheless to have evolved from the Cµ/ε 2 domain. • Hinge regions are encoded by separate exons. • The hinge region permits a considerable degree of flexibility between the antigen-binding and effector-interacting components of the immunoglobulin molecule.
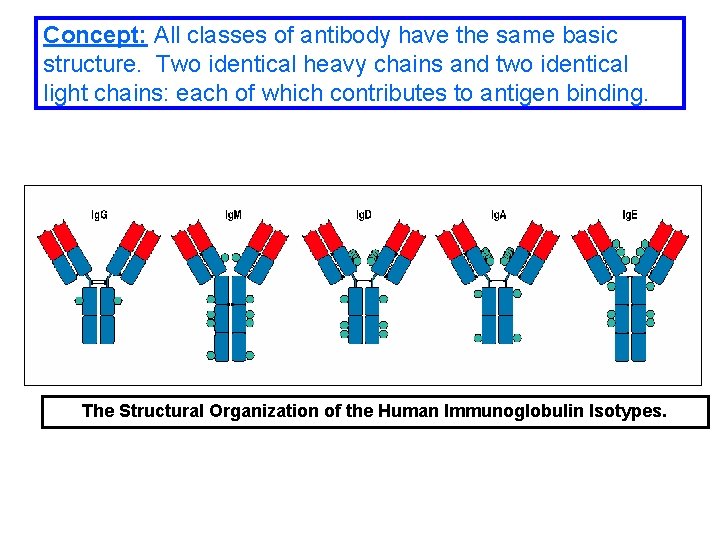
Concept: All classes of antibody have the same basic structure. Two identical heavy chains and two identical light chains: each of which contributes to antigen binding. The Structural Organization of the Human Immunoglobulin Isotypes.
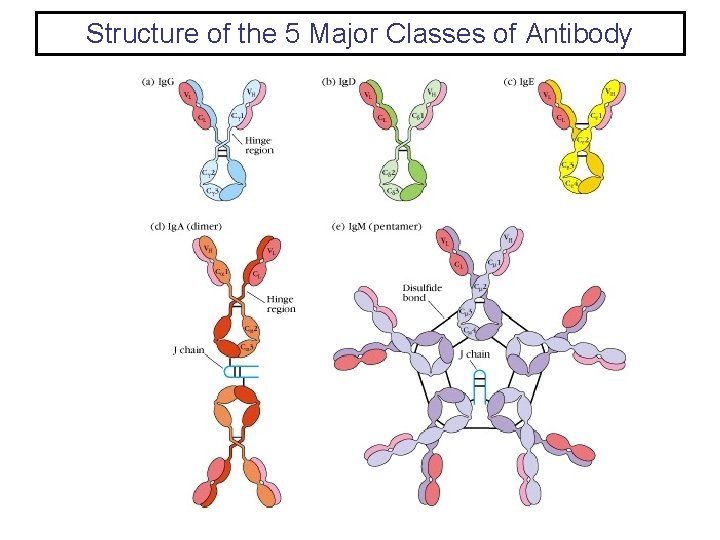
Structure of the 5 Major Classes of Antibody

Immunoglobulin G (Ig. G) • • The most abundant class in serum ~80% of the total serum immunoglobulins. Ig. G is found also in the interstitial spaces. The Ig. G molecule consists of two λ H-chains and two κ or two λ L-chains. In both mouse and man there are four subclasses of Ig. G fixes complement Ig. G is the only immunoglobulin to cross the placenta. Ig. G reacts with Fc. R’s on phagocytic cells to promote opsonization.
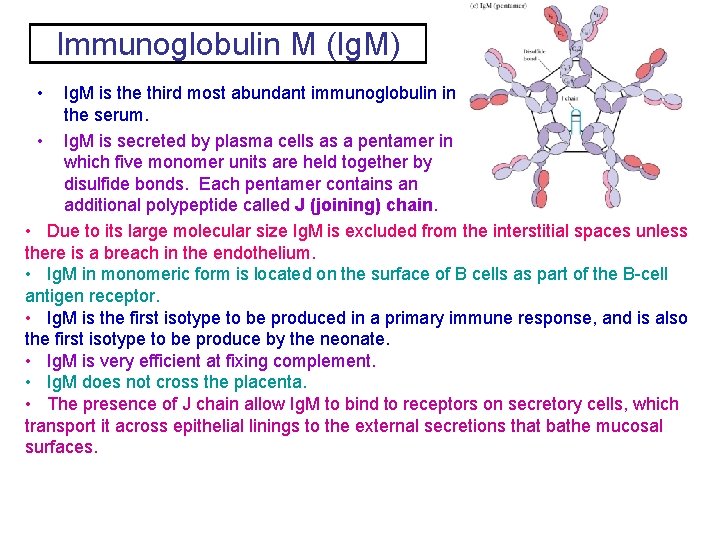
Immunoglobulin M (Ig. M) • Ig. M is the third most abundant immunoglobulin in the serum. • Ig. M is secreted by plasma cells as a pentamer in which five monomer units are held together by disulfide bonds. Each pentamer contains an additional polypeptide called J (joining) chain. • Due to its large molecular size Ig. M is excluded from the interstitial spaces unless there is a breach in the endothelium. • Ig. M in monomeric form is located on the surface of B cells as part of the B-cell antigen receptor. • Ig. M is the first isotype to be produced in a primary immune response, and is also the first isotype to be produce by the neonate. • Ig. M is very efficient at fixing complement. • Ig. M does not cross the placenta. • The presence of J chain allow Ig. M to bind to receptors on secretory cells, which transport it across epithelial linings to the external secretions that bathe mucosal surfaces.
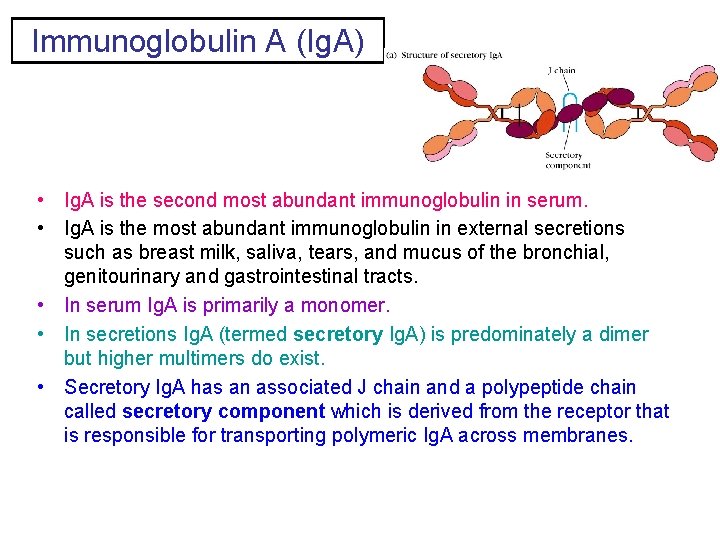
Immunoglobulin A (Ig. A) • Ig. A is the second most abundant immunoglobulin in serum. • Ig. A is the most abundant immunoglobulin in external secretions such as breast milk, saliva, tears, and mucus of the bronchial, genitourinary and gastrointestinal tracts. • In serum Ig. A is primarily a monomer. • In secretions Ig. A (termed secretory Ig. A) is predominately a dimer but higher multimers do exist. • Secretory Ig. A has an associated J chain and a polypeptide chain called secretory component which is derived from the receptor that is responsible for transporting polymeric Ig. A across membranes.
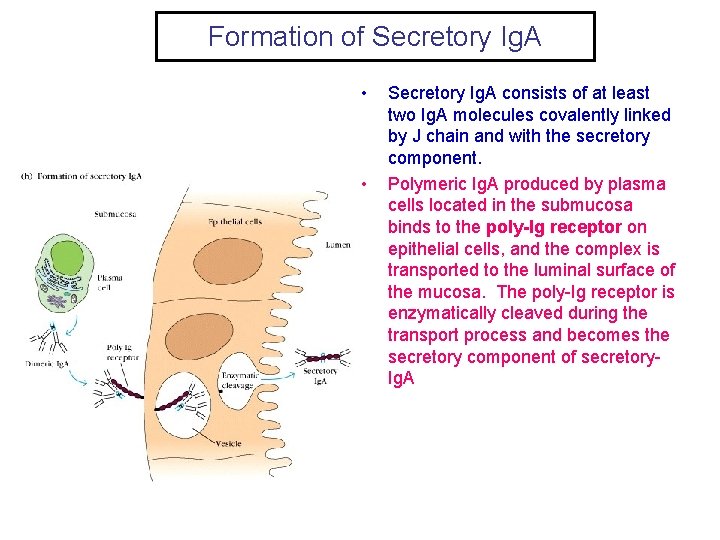
Formation of Secretory Ig. A • • Secretory Ig. A consists of at least two Ig. A molecules covalently linked by J chain and with the secretory component. Polymeric Ig. A produced by plasma cells located in the submucosa binds to the poly-Ig receptor on epithelial cells, and the complex is transported to the luminal surface of the mucosa. The poly-Ig receptor is enzymatically cleaved during the transport process and becomes the secretory component of secretory. Ig. A

Ig. D and Ig. E • The principal role of Ig. D: Located on the surface of B-lymphocytes and along with Ig. M serves the function of antigen recognition by the B cell. • There are no known effector functions of Ig. D • Ig. E binds to mast cells and basophils through a high affinity Fc. R and is involved in mediating hypersensitivity reactions. This will be the subject of a late lecture.
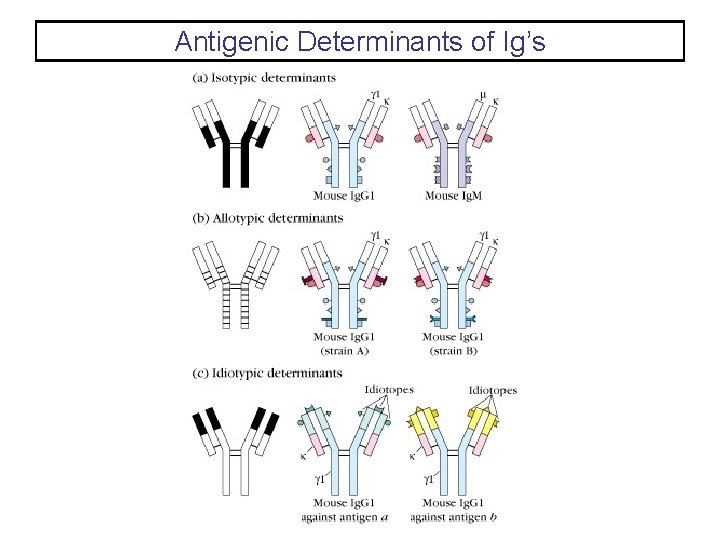
Antigenic Determinants of Ig’s
Antigenic Determinants on Ig’s • The antigenic determinants, or epitopes, on immunoglobulins fall into three major categories: – Genetic polymorphisms in antibody molecules are called allotypes. – The isotype of immunoglobulins is determined by the heavy chain. Humans have 9 isotypes of immunoglobulins. – The unique antigenic structures formed by the variable regions of immunoglobulins produces unique epitopes for each antibody specificity. These epitopes are called idiotopes. The collection of idiotopes define the idiotype of the immunoglobulin.
The Immunoglobulin Superfamily of Proteins • As you will learn throughout this course, many other proteins (including TCR, CD 3, MHC, CD 4, CD 8, Fc. R, ICAM 1…. etc. ) on the surface of lymphocytes and other cells, many of which function as cell-cell adhesion molecules, contain the characteristic immunoglobulin fold. • This observation suggests that the genes encoding these proteins have a common evolutionary history. • Most of the amino acids in each Ig-like domain are usually encoded by a single exon and it seems likely that the entire supergene family evolved from a gene coding for a single Ig-like domain that may have been involved in cell-cell interactions. • Evidence suggests that the primordial gene arose before vertebrates diverged from invertebrate ancestor some 400 million years ago. • New family members presumably arose by exon and gene duplications, events that probably gave rise to the multiple gene segments that encode antibodies and T cell receptors.
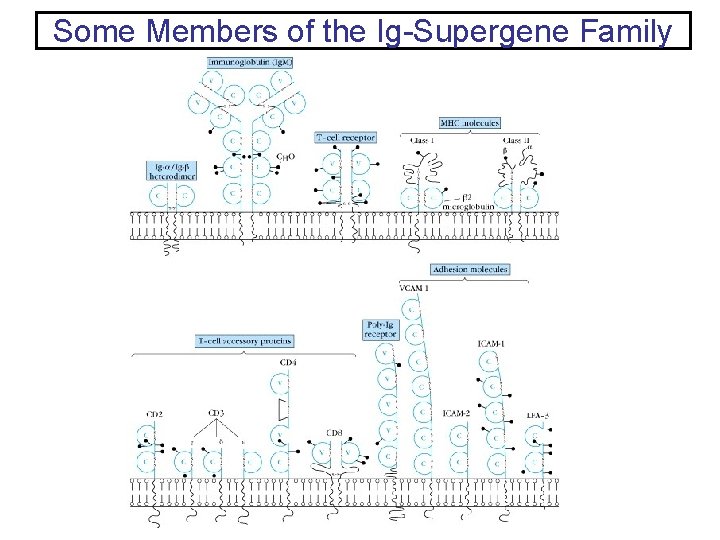
Some Members of the Ig-Supergene Family
Generation of Lymphocyte Antigen Receptor • The expression of the antigen receptor is the defining (and essential) event in the development of both B and T cells. • Antigen receptors, in the form of Ig’s on B cells and the TCR on T-cells, are the means by which lymphocytes sense the presence of antigen in their environment. • The diverse repertoire of lymphocyte receptors is accomplished through complex and elegant genetic mechanisms. • The basic mechanism for generation of diversity is common to both B cells and T cells and involves many if not all of the same enzymes.
Recall these Facts • The receptors produced by each lymphocyte have a unique antigen specificity which is determined by the structure of their antigen-binding site. • The wide range of antigen specificities in the antigen receptor repertoire is due to variation in the amino acid sequence in the V region. • Each individual possesses billions of lymphocytes, these cells collectively provide the individual with the ability to respond to a great variety of antigens. • In each chain the V region is linked to an invariant constant region which provides effector function
Gene Segments of VL and VH Regions • Light chain V region – two gene segments – V gene segment –first 95 -101 amino acids – J (joining) gene segment – up to 13 amino acids • Heavy chain V region – three gene segments – V and J gene segments – D (diversity) gene segment
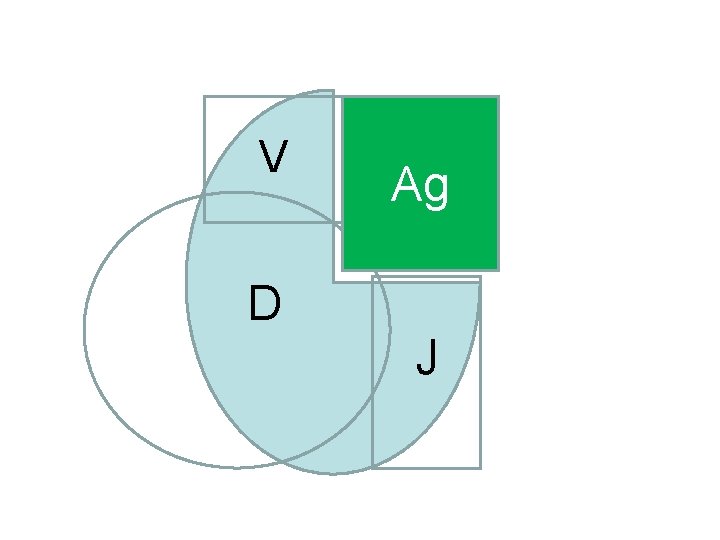
V Ag D J
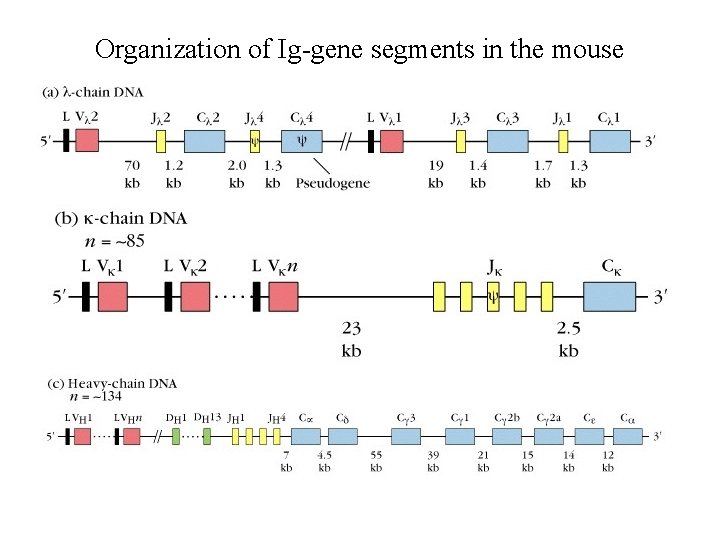
Organization of Ig-gene segments in the mouse
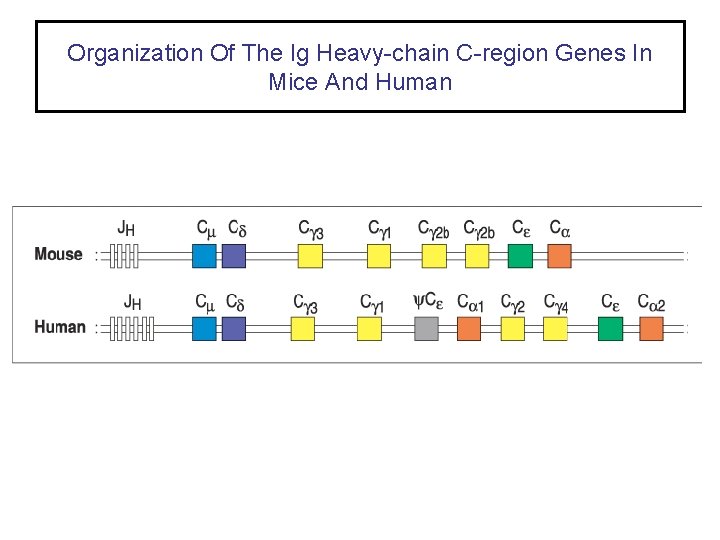
Organization Of The Ig Heavy-chain C-region Genes In Mice And Human
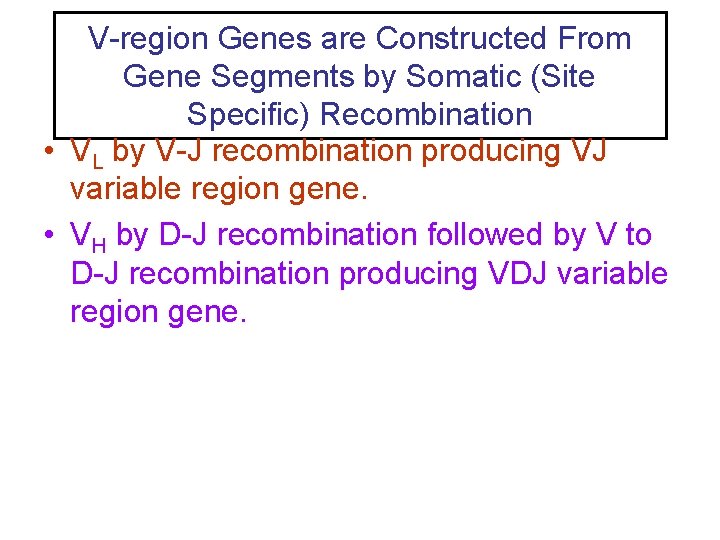
V-region Genes are Constructed From Gene Segments by Somatic (Site Specific) Recombination • VL by V-J recombination producing VJ variable region gene. • VH by D-J recombination followed by V to D-J recombination producing VDJ variable region gene.
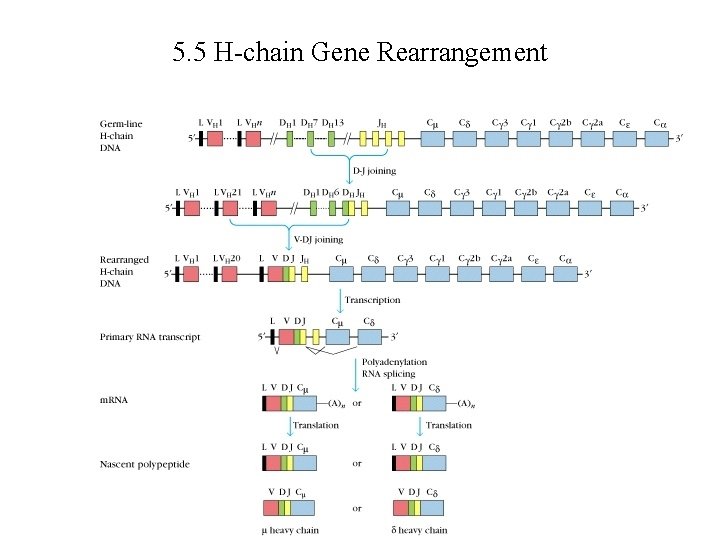
5. 5 H-chain Gene Rearrangement
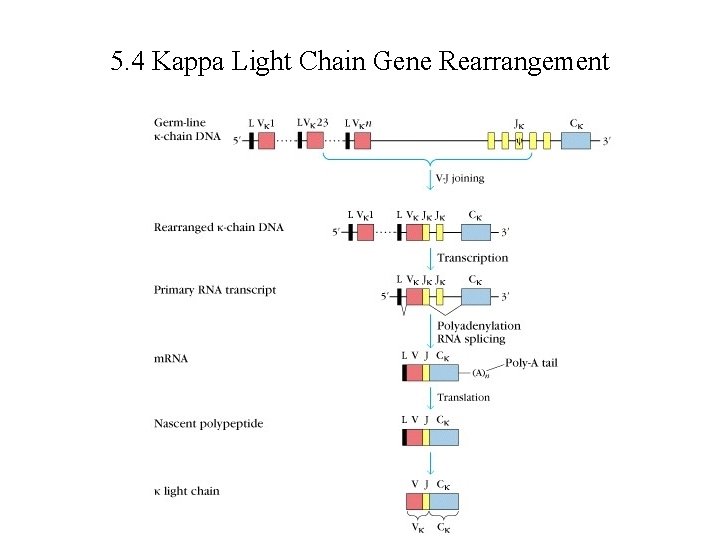
5. 4 Kappa Light Chain Gene Rearrangement
Structural Variation in Immunoglobulin Constant Regions • The immunoglobulin H-chain isotype are distinguished by the structure of their constant regions. • Antibody C-regions confer functional specialization. • Co-expression of Ig. M and Ig. D on B cells results from alternatively spliced H-chain transcripts. • Transmembrane and secreted forms of Ig’s are generated from alternative H-chain transcripts. • CLASS SWITCHING – the same VH exon can associate with different CH genes in the course of an immune response.
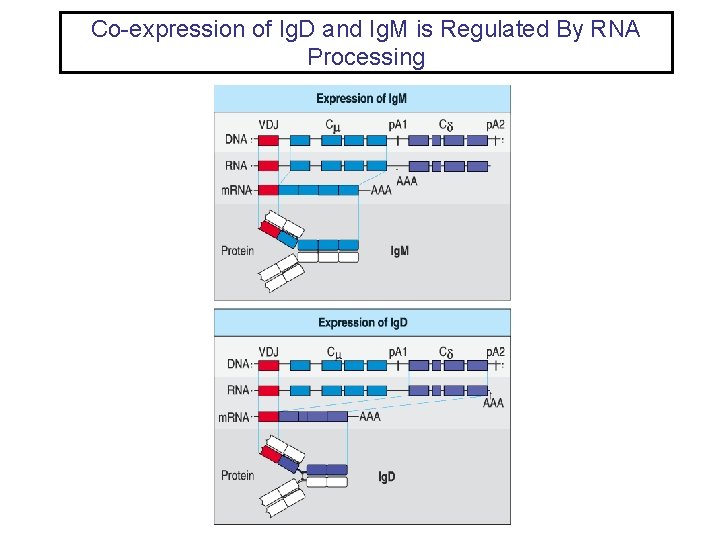
Co-expression of Ig. D and Ig. M is Regulated By RNA Processing
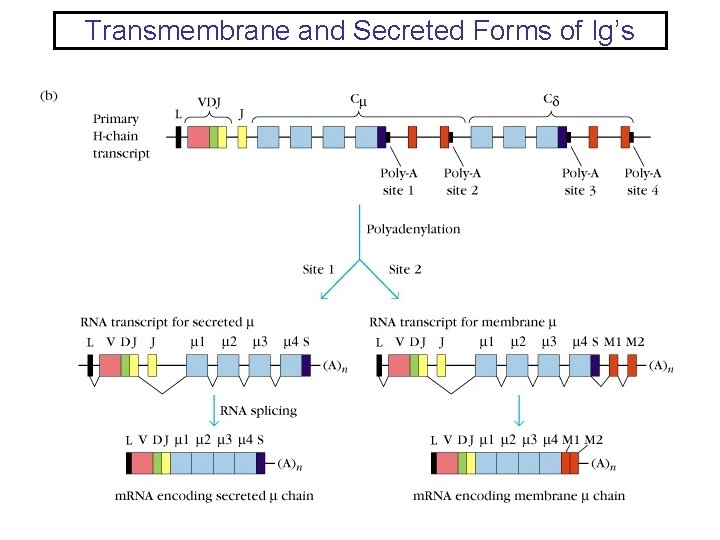
Transmembrane and Secreted Forms of Ig’s
Transmembrane and Secreted Forms of Ig’s Both forms are derived from the same H-chain gene sequence • Each H-chain has: – Two exons that encode the transmembrane region and the cytoplasmic tail – One exon that encodes the carboxy-terminus of the secreted form • The events that dictate whether a H-chain RNA will result in secreted or transmembrane occur during processing of the initial transcript. • The selection of transmembrane or secreted form is developmentally regulated. Prior to antigen stimualtion B cells make predominately the transmembrane form. However, plasma cell make exclusively the secreted form.
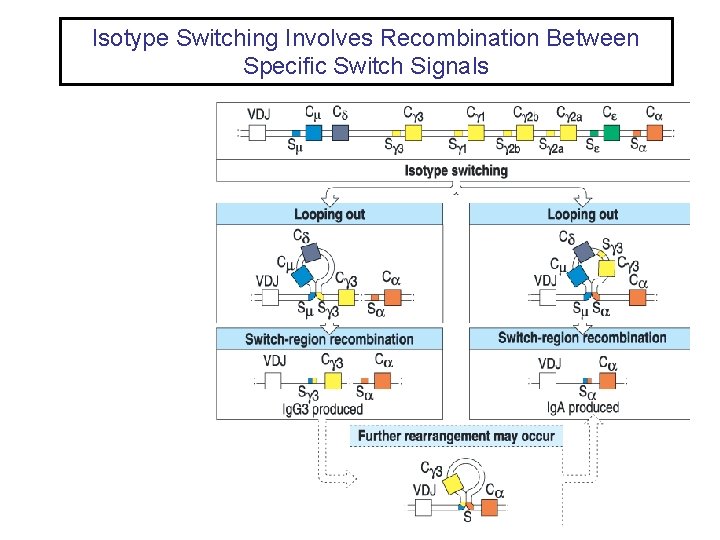
Isotype Switching Involves Recombination Between Specific Switch Signals
Isotype Switching • Repetitive DNA sequences that guide isotype switching are found upstream of each of the C-region genes. • Switching occurs by recombination between these repetitive sequences (switch signals). • Isotype switching results in deletion of the intervening DNA • Since the intervening DNA is deleted back switches are not possible, but additional switches to down stream isotypes is possible • The initial switching event takes place from the µ switch region • Subsequent switches to other isotypes take place from the recombinant switch region formed after µ switching. • Isotype switching is unlike V(D)J recombination is several ways – – All isotype switching is productive It uses different recombination signal sequences and enzymes It happens after antigen stimulation not during B cell development The switching process is not random – it is regulated by external signals from T cells

Somatic Hypermutation Somatic hypermutation further diversifies the Ab repertoire • Introduces variation into the rearranged immunoglobulin V-region that is subject to positive and negative selection • Occurs in the germinal center following antigen stimulation of the B cell. • Somatic hypermutation requires signals from activated T cells • Hypermutation is thought to occur due to the introduction of double strand breaks in the DNA of V regions, followed by error prone repair. • Hypermutation occurs at a similar time to class switching, but appear to involve different enzymes and mechanisms. • We will cover hypermutation in more detail in the lectures on B cell activation.
Regulation of Ig-Gene Transcription • Immunoglobulin genes are expressed only in B cells • Genes are expressed at different rates during different stages of development • Three major classes of cis regulatory sequences in DNA regulate transcription of Ig-genes – Promoters – short nucleotide sequences extending about 200 bp upstream from the start site, that promote intiation of RNA transcription – orientation dependent. – Ehancers – nucleotide sequences siturate some distance upstream or downstream from a gene that activate transcription from the promoter sequence in an orientationindependent manner – Silencers – nucleotide sequences that down-regulate transcription, operating in both directions over a distance
Generation of Antibody Diversity 1. 2. 3. 4. 5. 6. Multiple germline gene segments Combinatorial V-(D)-J joining Junctional flexibility P-region nucleotide addition (P-addition) N-region nucleotide addition (N-addition) Combinatorial association of light and heavy chains. 7. Somatic hypermutation
Summary: The combination of many sources of diversity generates a vast repertoire of antibody specificities from a limited number of genes Diversity with in the Ig repertoire is achieved by several means. 1. V regions are encoded by separate gene segments, which can be brought together by somatic recombination to make a complete V region gene. 2. Many V region gene segments are present in the genome, thus providing a heritable source of diversity. 3. Combinatorial diversity results from the random recombination of separate V , D and J gene segments to form a complete V region exon. 4. Variability at the joints is increased by N-region and P-region additions and by the variable deletion of nucleotides at the ends of coding sequences. 5. The association if different light and heavy chain V regions to form the antigen-binding site of an Ig molecule contributes further to the diversity. 6. Finally, after an immunoglobulin is expressed, the coding regions of the V regions are modified by somatic hypermutation following stimulation of the B cell by antigen.
- Slides: 55