Annotation of eukaryotic genomes Genomic DNA ab initio
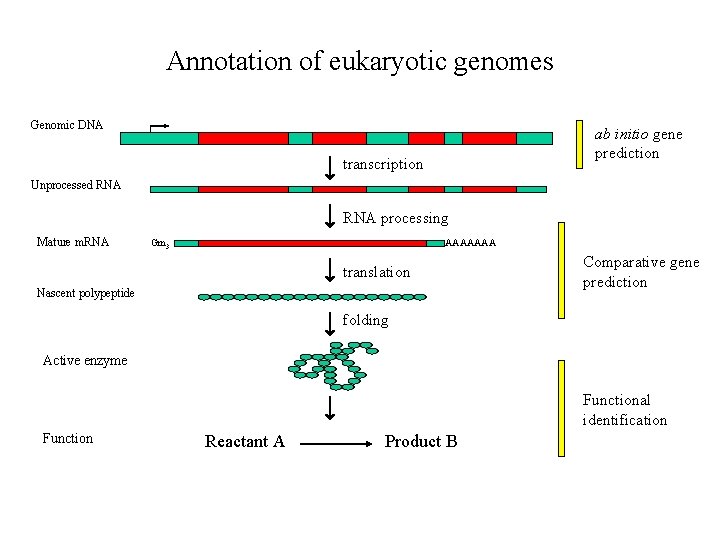
Annotation of eukaryotic genomes Genomic DNA ab initio gene prediction transcription Unprocessed RNA processing Mature m. RNA Gm 3 AAAAAAA translation Nascent polypeptide Comparative gene prediction folding Active enzyme Functional identification Function Reactant A Product B
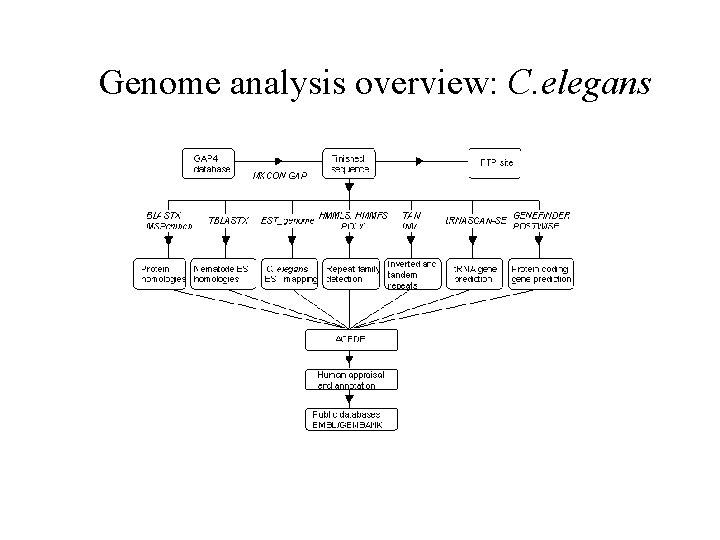
Genome analysis overview: C. elegans

Gene finding: ab initio • What features of a ORF can we use? • Size - large open reading frames • DNA composition - codon usage / 3 rd position codon bias • Other features: • Kozak sequence CCGCCAUGG • Ribosome binding sites • Termination signal (stops) • Splice junction boundaries

Gene finding: comparative • Use knowledge of known coding sequences to identify region of genomic DNA by similarity • transcribed DNA sequence • peptide sequence • related genomic sequence
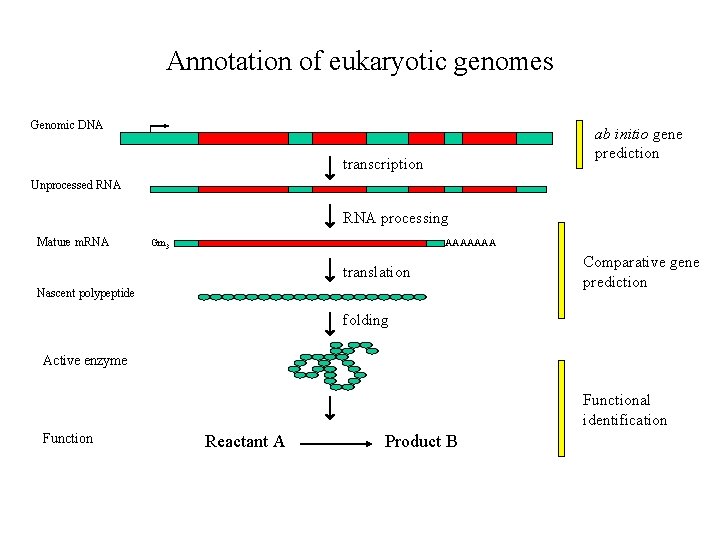
Annotation of eukaryotic genomes Genomic DNA ab initio gene prediction transcription Unprocessed RNA processing Mature m. RNA Gm 3 AAAAAAA translation Nascent polypeptide Comparative gene prediction folding Active enzyme Functional identification Function Reactant A Product B
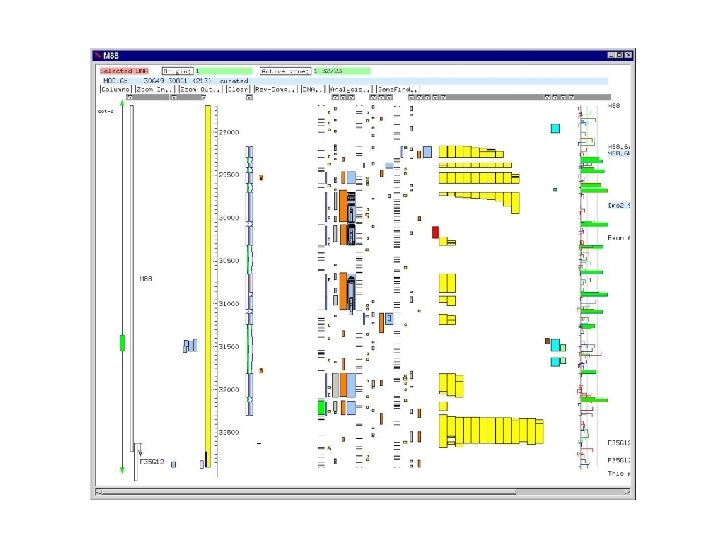
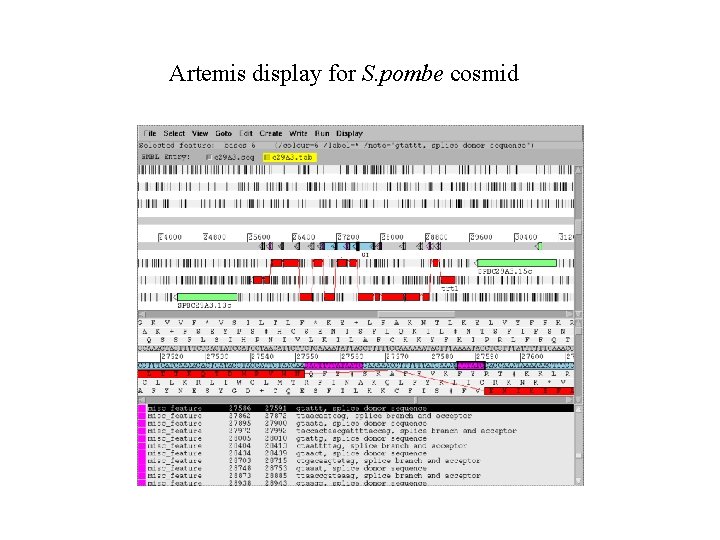
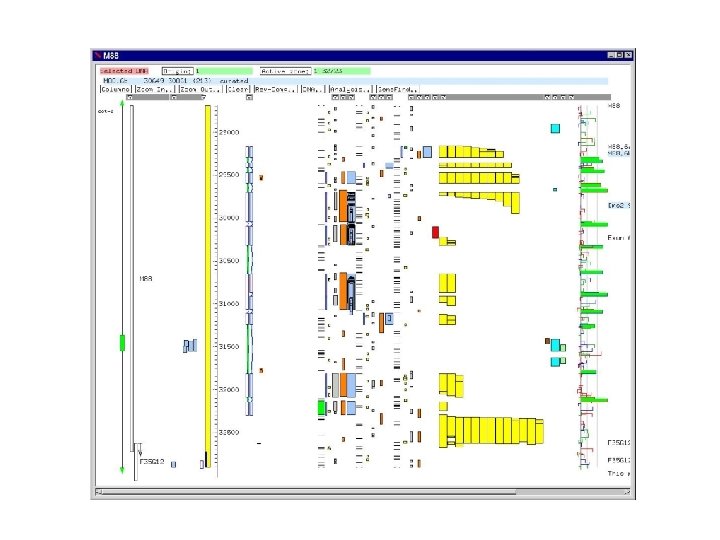
Artemis display for S. pombe cosmid

Methods for searching • Pairwise alignments: matching a query sequence against a database of subject sequences • Needleman & Wunsch - global alignment • Smith-Waterman - local alignment • Fast. A • BLAST • Others: SSAHA, WABA • see Chapter 7 Developing Bionformatics Computer Skills
BLAST - local similarity searches • BLAST (Basic Local Alignment Search Tool) is the workhorse of genome annotation due to it’s early optimisation for the UNIX platform • Underlies most of the web-based servers world-wide • Comes in many flavours: • BLASTN - DNA against DNA • BLASTX - DNA against Protein • BLASTP - Protein against Protein • TBLASTN - Protein against DNA • TBLASTX - DNA against DNA at the peptide level

BLAST - results • BLAST returns high-scoring pairs (HSPs) with a score and p-value. Blast output files can be large and difficult to interpret. • Hence we need tools to make sense of the data - both to filter/process the file and to visualise the resulting multiple sequence alignments. • MSPcrunch - a post-processor for BLAST with a number of different output types. • Bio. Perl BLAST output - modules for handling sequences and
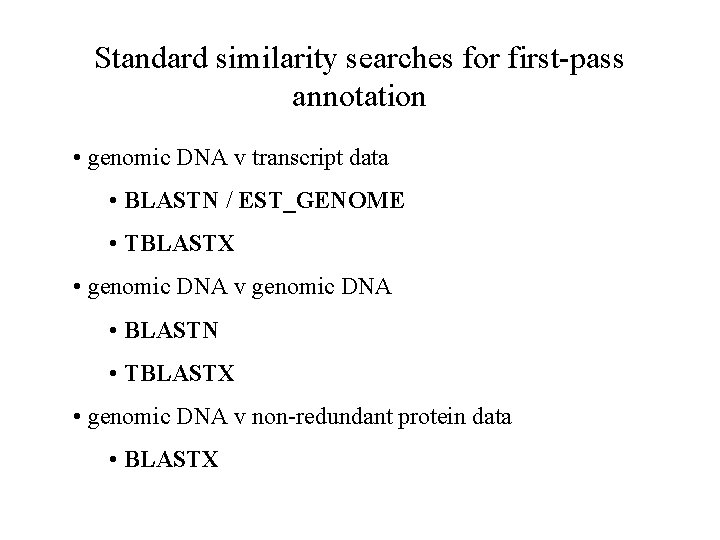
Standard similarity searches for first-pass annotation • genomic DNA v transcript data • BLASTN / EST_GENOME • TBLASTX • genomic DNA v genomic DNA • BLASTN • TBLASTX • genomic DNA v non-redundant protein data • BLASTX
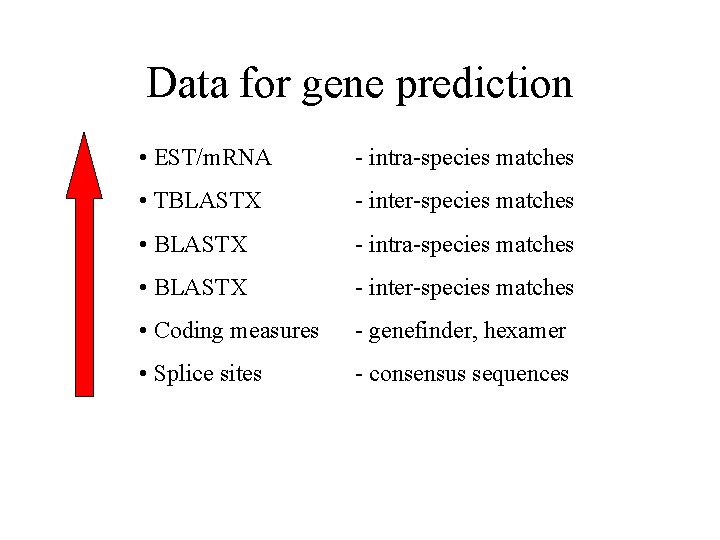
Data for gene prediction • EST/m. RNA - intra-species matches • TBLASTX - inter-species matches • BLASTX - intra-species matches • BLASTX - inter-species matches • Coding measures - genefinder, hexamer • Splice sites - consensus sequences
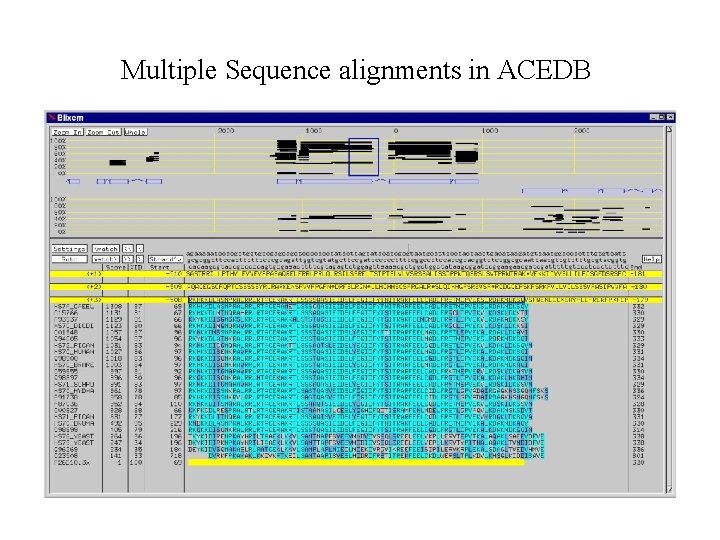
Multiple Sequence alignments in ACEDB
Manual review of gene predictions • Check concordance with transcript data • Check concordance with peptide similarity data • Check splice site usage (intron / exon boundaries) • Set of human appraised gene predictions. The translations of the CDS sequences are used for protein feature analysis and initial assignment (ID, function)

- Slides: 18