Analyzing SmokeExposed Wine Grapes Applying Data Reduction and


- Slides: 13

Analyzing Smoke-Exposed Wine Grapes: Applying Data Reduction and Data Management Tools DATA 501 Project Presentation Thursday, March 30, 2017

Wine Flavour 2

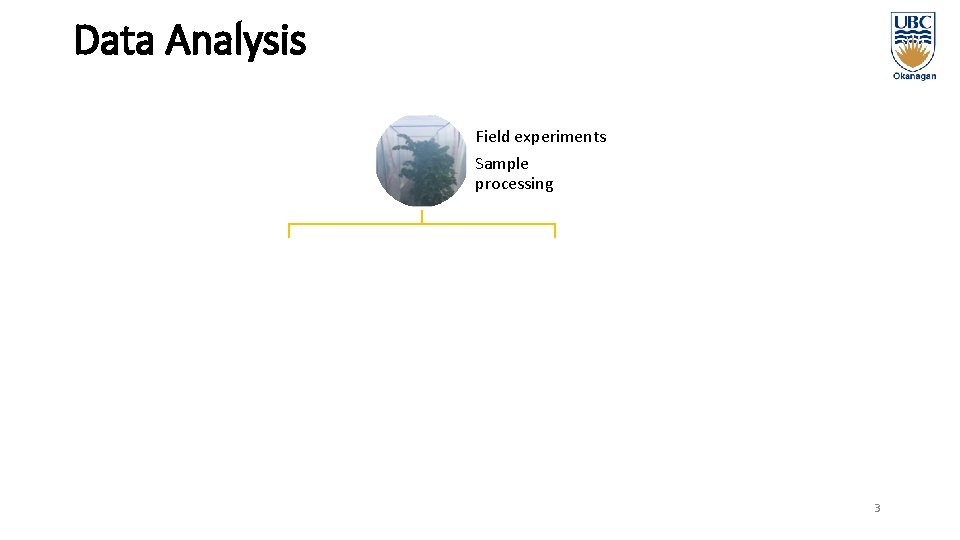
Data Analysis Non-Targeted Analysis Field experiments Sample processing a a Compound identification Quantitative analysis a a Compound significance Summary statistics and Comparisons 3

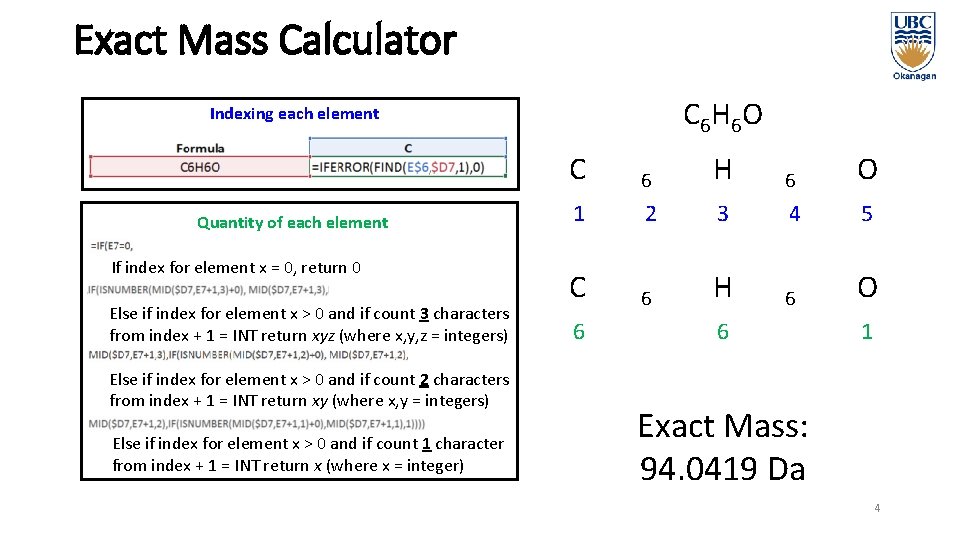
Exact Mass Calculator C 6 H 6 O Indexing each element Quantity of each element If index for element x = 0, return 0 Else if index for element x > 0 and if count 3 characters from index + 1 = INT return xyz (where x, y, z = integers) Else if index for element x > 0 and if count 2 characters from index + 1 = INT return xy (where x, y = integers) Else if index for element x > 0 and if count 1 character from index + 1 = INT return x (where x = integer) C 6 H 6 O 1 2 3 4 5 C 6 H 6 O 6 6 1 Exact Mass: 94. 0419 Da 4

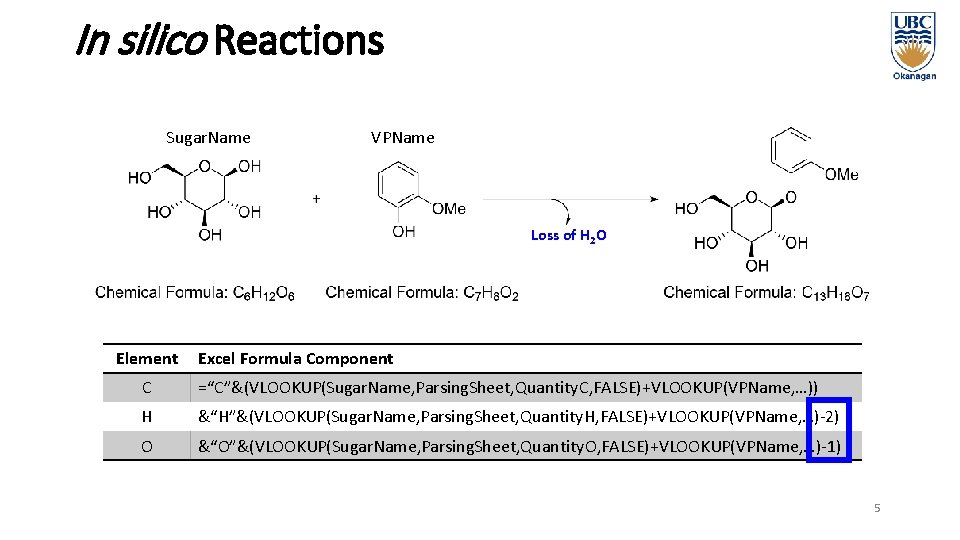
In silico Reactions Sugar. Name VPName Loss of H 2 O Element Excel Formula Component C =“C”&(VLOOKUP(Sugar. Name, Parsing. Sheet, Quantity. C, FALSE)+VLOOKUP(VPName, …)) H &“H”&(VLOOKUP(Sugar. Name, Parsing. Sheet, Quantity. H, FALSE)+VLOOKUP(VPName, …)-2) O &“O”&(VLOOKUP(Sugar. Name, Parsing. Sheet, Quantity. O, FALSE)+VLOOKUP(VPName, …)-1) 5

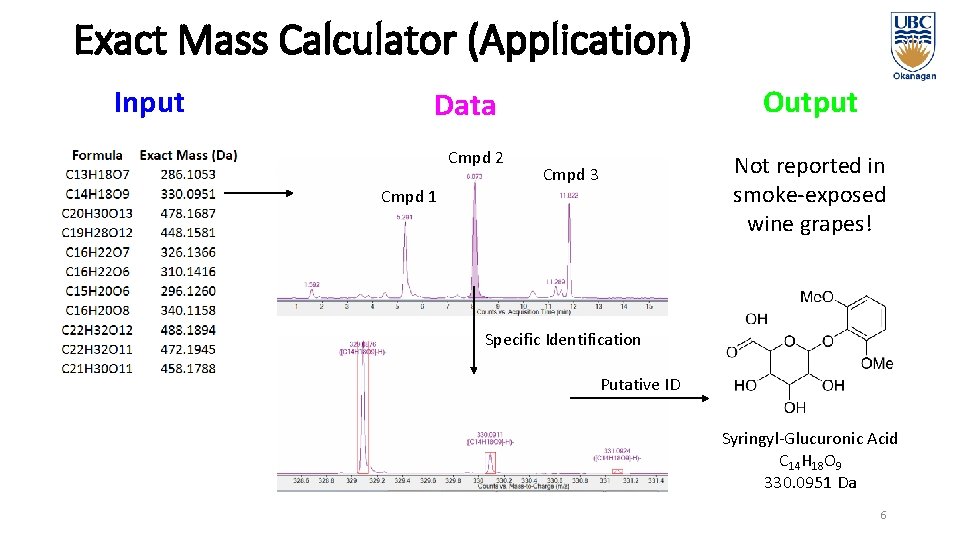
Exact Mass Calculator (Application) Input Output Data Cmpd 2 Not reported in smoke-exposed wine grapes! Cmpd 3 Cmpd 1 Specific Identification Putative ID Syringyl-Glucuronic Acid C 14 H 18 O 9 330. 0951 Da 6

Relational Databases ~10, 000 quantitative values to process + Undefined qualitative values 7

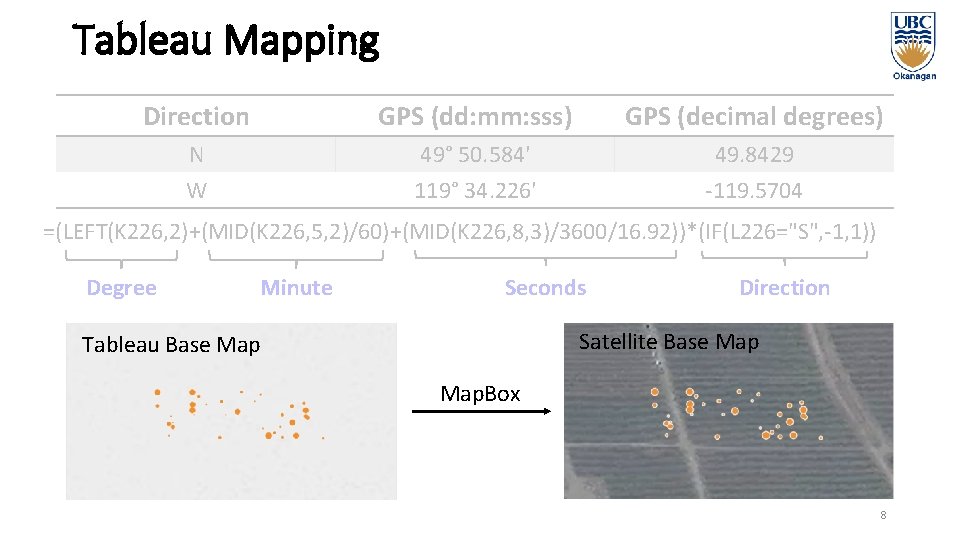
Tableau Mapping Direction GPS (dd: mm: sss) GPS (decimal degrees) N W 49° 50. 584' 119° 34. 226' 49. 8429 -119. 5704 =(LEFT(K 226, 2)+(MID(K 226, 5, 2)/60)+(MID(K 226, 8, 3)/3600/16. 92))*(IF(L 226="S", -1, 1)) Degree Minute Seconds Direction Satellite Base Map Tableau Base Map. Box 8

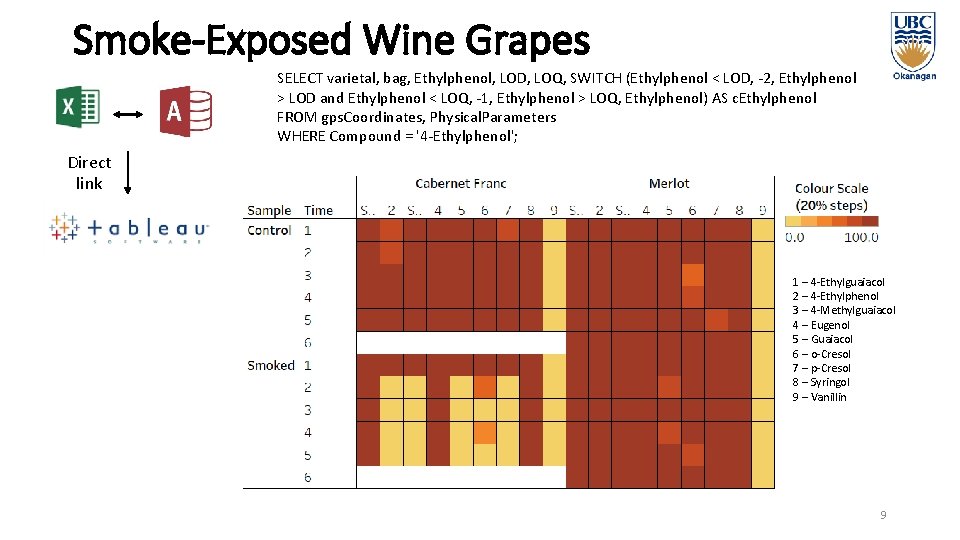
Smoke-Exposed Wine Grapes SELECT varietal, bag, Ethylphenol, LOD, LOQ, SWITCH (Ethylphenol < LOD, -2, Ethylphenol > LOD and Ethylphenol < LOQ, -1, Ethylphenol > LOQ, Ethylphenol) AS c. Ethylphenol FROM gps. Coordinates, Physical. Parameters WHERE Compound = '4 -Ethylphenol'; Direct link 1 – 4 -Ethylguaiacol 2 – 4 -Ethylphenol 3 – 4 -Methylguaiacol 4 – Eugenol 5 – Guaiacol 6 – o-Cresol 7 – p-Cresol 8 – Syringol 9 – Vanillin 9

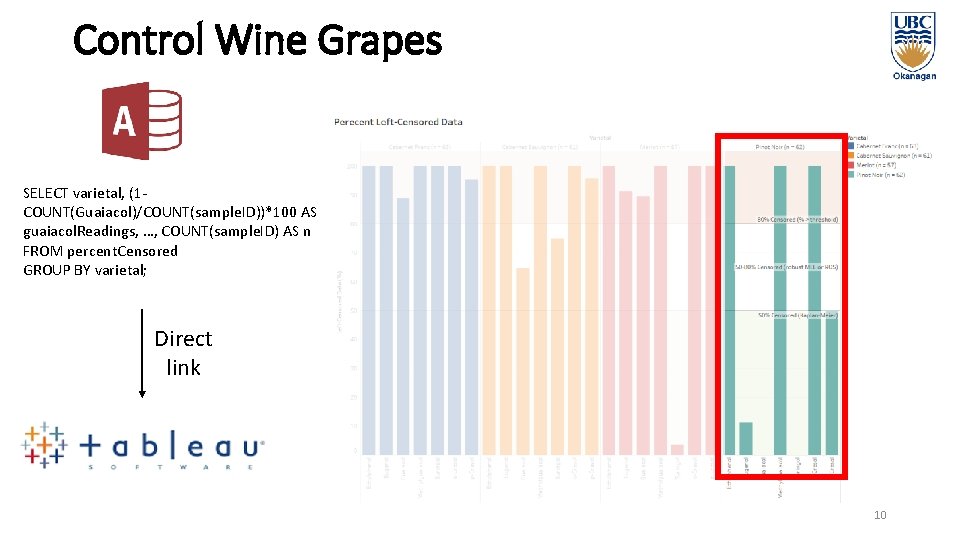
Control Wine Grapes SELECT varietal, (1 COUNT(Guaiacol)/COUNT(sample. ID))*100 AS guaiacol. Readings, …, COUNT(sample. ID) AS n FROM percent. Censored GROUP BY varietal; Direct link 10

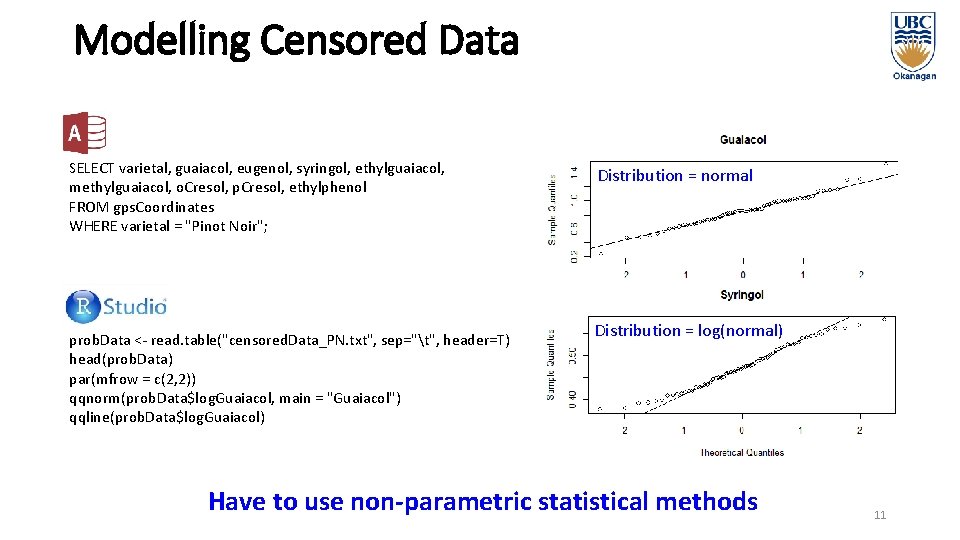
Modelling Censored Data SELECT varietal, guaiacol, eugenol, syringol, ethylguaiacol, methylguaiacol, o. Cresol, p. Cresol, ethylphenol FROM gps. Coordinates WHERE varietal = "Pinot Noir"; prob. Data <- read. table("censored. Data_PN. txt", sep="t", header=T) head(prob. Data) par(mfrow = c(2, 2)) qqnorm(prob. Data$log. Guaiacol, main = "Guaiacol") qqline(prob. Data$log. Guaiacol) Distribution = normal Distribution = log(normal) Have to use non-parametric statistical methods 11

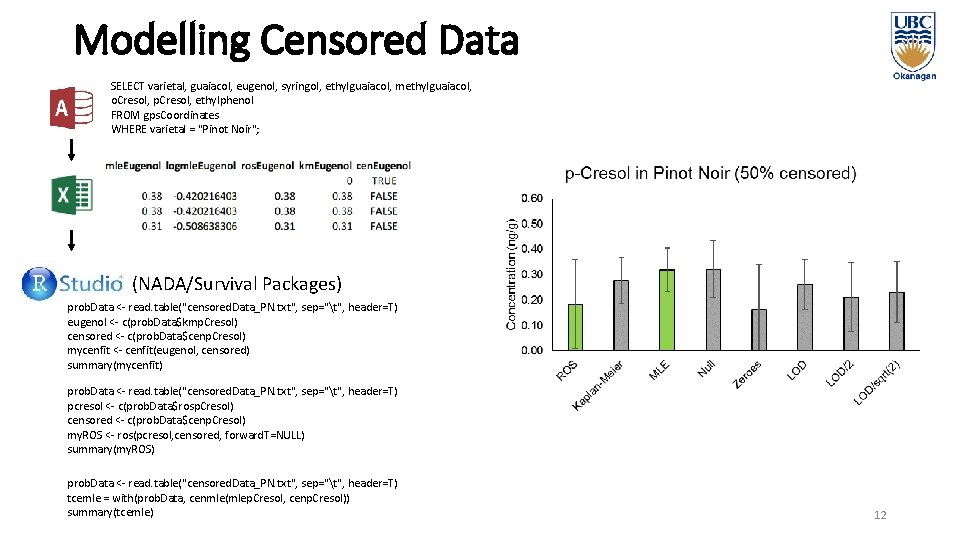
Modelling Censored Data SELECT varietal, guaiacol, eugenol, syringol, ethylguaiacol, methylguaiacol, o. Cresol, p. Cresol, ethylphenol FROM gps. Coordinates WHERE varietal = "Pinot Noir"; (NADA/Survival Packages) prob. Data <- read. table("censored. Data_PN. txt", sep="t", header=T) eugenol <- c(prob. Data$kmp. Cresol) censored <- c(prob. Data$cenp. Cresol) mycenfit <- cenfit(eugenol, censored) summary(mycenfit) prob. Data <- read. table("censored. Data_PN. txt", sep="t", header=T) pcresol <- c(prob. Data$rosp. Cresol) censored <- c(prob. Data$cenp. Cresol) my. ROS <- ros(pcresol, censored, forward. T=NULL) summary(my. ROS) prob. Data <- read. table("censored. Data_PN. txt", sep="t", header=T) tcemle = with(prob. Data, cenmle(mlep. Cresol, cenp. Cresol)) summary(tcemle) 12

Summary and Outlook My Results Unreported compound in smoke-exposed wine grapes Ongoing Automate hypothesis testing in R Adjustments to field-based smoking procedures Guidance for statistical calculations 13