Analysis Tools in Panda Root Glue X PANDA
















![Filter Syntax: • Counts: "(<particle> ; [<mult>] ; [kinematic cuts])" <particle>: particle type (MC Filter Syntax: • Counts: "(<particle> ; [<mult>] ; [kinematic cuts])" <particle>: particle type (MC](https://slidetodoc.com/presentation_image_h2/5a413baab1f8a3cddf628100ce512ee5/image-92.jpg)
- Slides: 94

Analysis Tools in Panda. Root Glue. X PANDA Workshop 2019 Washington, GW, May 3 - 5, 2019 Klaus Götzen GSI Darmstadt

Topics • Performing analysis – The Rho Analysis Framework – Panda. Root Tools Only briefly discussed • Previewing signal/background reconstruction – Fast Simulations • Studying generic signal/background distributions – Event Generators - Stand-Alone Usage • Reduce computing demand (background simulation) – Event Filtering • Special Feature: – Speeding up Glue. X interactive analysis K. Götzen Glue. X PANDA Workshop 2019 2

Performing Analysis

The Rho Analysis Framework • Core based on the Rho Analysis Framework – Inspired by the beta software/analysis framework of Ba. Bar – Written by M. Kunze at Univ. Bochum – Fully ROOT based – Optimized for speed • Panda. ROOT Analysis Toolset – Analysis via ROOT Macros – Extends core by experiment specific features – PID, MC Truth match – Simple Analysis Tools – QA Tools K. Götzen Glue. X PANDA Workshop 2019 4

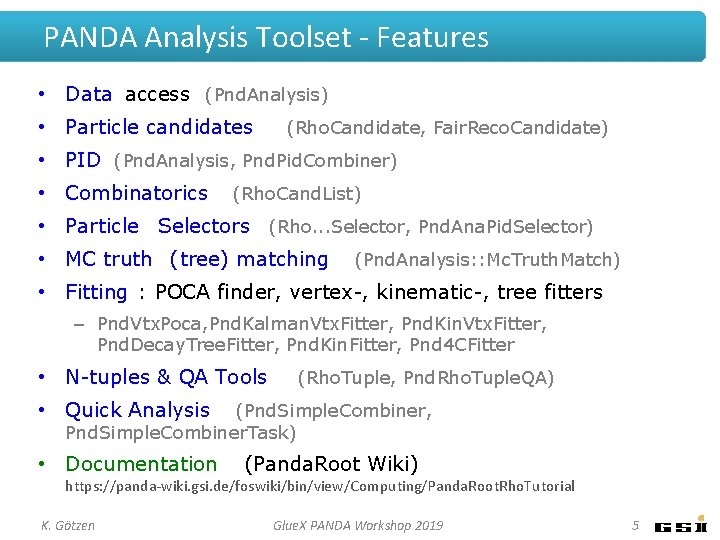
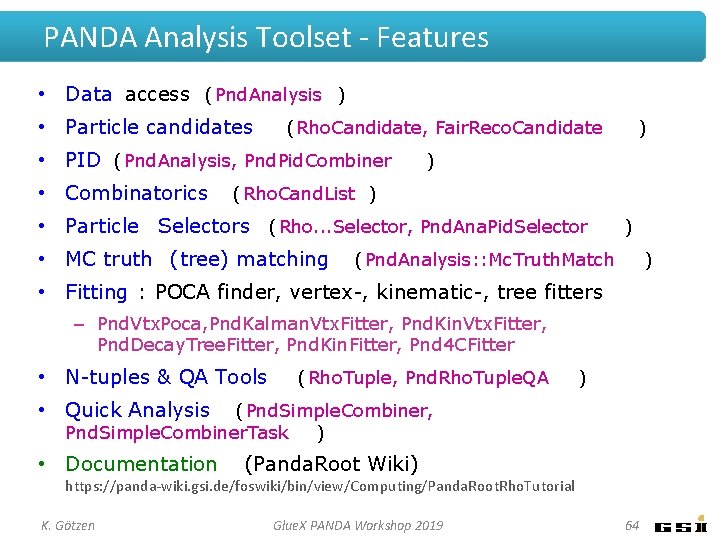
PANDA Analysis Toolset - Features • Data access (Pnd. Analysis) • Particle candidates (Rho. Candidate, Fair. Reco. Candidate) • PID (Pnd. Analysis, Pnd. Pid. Combiner) • Combinatorics (Rho. Cand. List) • Particle Selectors (Rho. . . Selector, Pnd. Ana. Pid. Selector) • MC truth (tree) matching (Pnd. Analysis: : Mc. Truth. Match) • Fitting : POCA finder, vertex-, kinematic-, tree fitters – Pnd. Vtx. Poca, Pnd. Kalman. Vtx. Fitter, Pnd. Kin. Vtx. Fitter, Pnd. Decay. Tree. Fitter, Pnd. Kin. Fitter, Pnd 4 CFitter • N-tuples & QA Tools (Rho. Tuple, Pnd. Rho. Tuple. QA) • Quick Analysis (Pnd. Simple. Combiner, Pnd. Simple. Combiner. Task) • Documentation (Panda. Root Wiki) https: //panda-wiki. gsi. de/foswiki/bin/view/Computing/Panda. Root. Rho. Tutorial K. Götzen Glue. X PANDA Workshop 2019 5

Basic Analysis

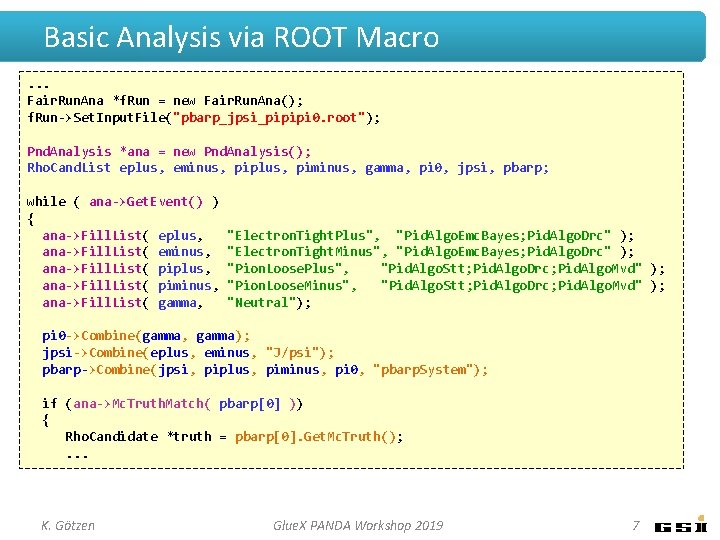
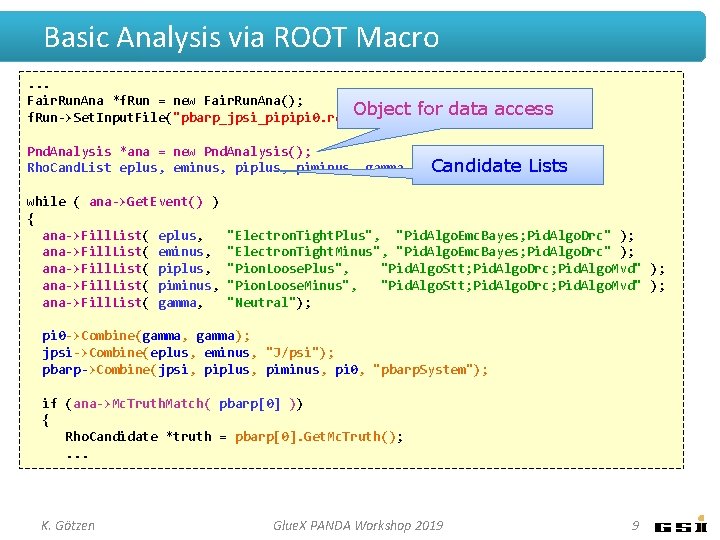
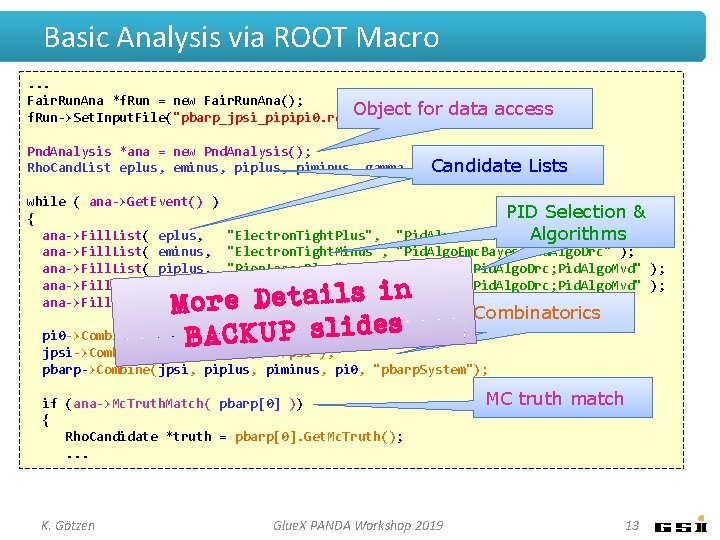
Basic Analysis via ROOT Macro. . . Fair. Run. Ana *f. Run = new Fair. Run. Ana(); f. Run->Set. Input. File("pbarp_jpsi_pipipi 0. root"); Pnd. Analysis *ana = new Pnd. Analysis(); Rho. Cand. List eplus, eminus, piplus, piminus, gamma, pi 0, jpsi, pbarp; while ( ana->Get. Event() ) { ana->Fill. List( eplus, ana->Fill. List( eminus, ana->Fill. List( piplus, ana->Fill. List( piminus, ana->Fill. List( gamma, "Electron. Tight. Plus", "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" ); "Electron. Tight. Minus", "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" ); "Pion. Loose. Plus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Pion. Loose. Minus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Neutral"); pi 0 ->Combine(gamma, gamma); jpsi->Combine(eplus, eminus, "J/psi"); pbarp->Combine(jpsi, piplus, piminus, pi 0, "pbarp. System"); if (ana->Mc. Truth. Match( pbarp[0] )) { Rho. Candidate *truth = pbarp[0]. Get. Mc. Truth(); . . . K. Götzen Glue. X PANDA Workshop 2019 7

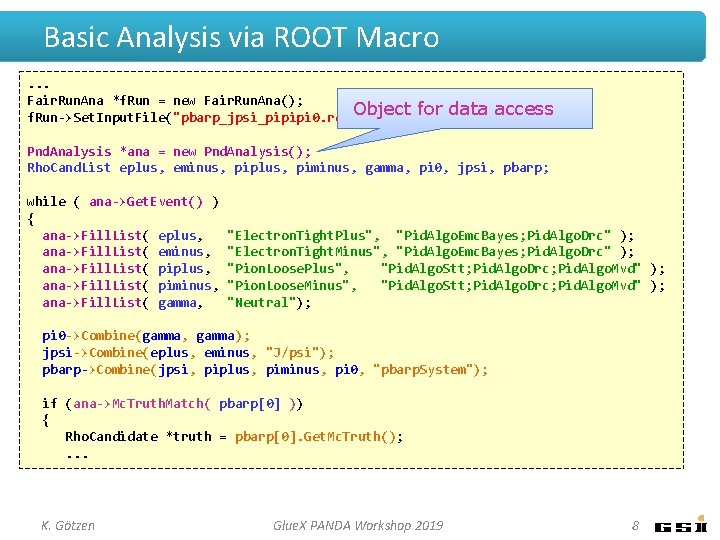
Basic Analysis via ROOT Macro. . . Fair. Run. Ana *f. Run = new Fair. Run. Ana(); Object f. Run->Set. Input. File("pbarp_jpsi_pipipi 0. root"); for data access Pnd. Analysis *ana = new Pnd. Analysis(); Rho. Cand. List eplus, eminus, piplus, piminus, gamma, pi 0, jpsi, pbarp; while ( ana->Get. Event() ) { ana->Fill. List( eplus, ana->Fill. List( eminus, ana->Fill. List( piplus, ana->Fill. List( piminus, ana->Fill. List( gamma, "Electron. Tight. Plus", "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" ); "Electron. Tight. Minus", "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" ); "Pion. Loose. Plus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Pion. Loose. Minus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Neutral"); pi 0 ->Combine(gamma, gamma); jpsi->Combine(eplus, eminus, "J/psi"); pbarp->Combine(jpsi, piplus, piminus, pi 0, "pbarp. System"); if (ana->Mc. Truth. Match( pbarp[0] )) { Rho. Candidate *truth = pbarp[0]. Get. Mc. Truth(); . . . K. Götzen Glue. X PANDA Workshop 2019 8

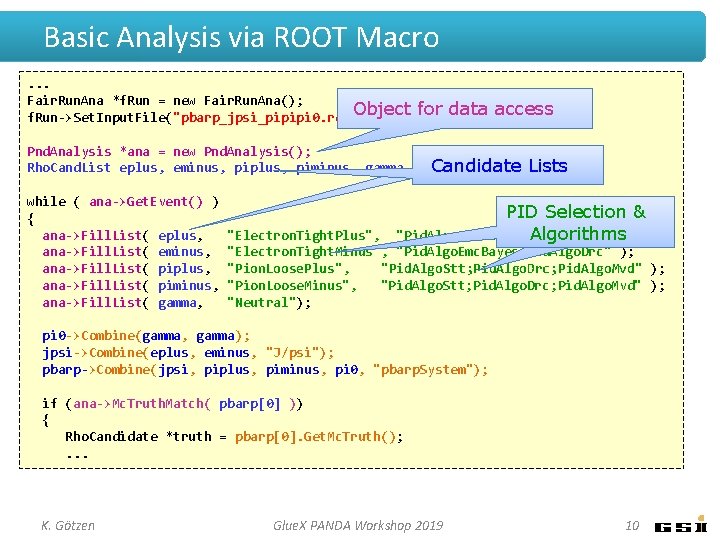
Basic Analysis via ROOT Macro. . . Fair. Run. Ana *f. Run = new Fair. Run. Ana(); Object f. Run->Set. Input. File("pbarp_jpsi_pipipi 0. root"); for data access Pnd. Analysis *ana = new Pnd. Analysis(); Candidate Lists Rho. Cand. List eplus, eminus, piplus, piminus, gamma, pi 0, jpsi, pbarp; while ( ana->Get. Event() ) { ana->Fill. List( eplus, ana->Fill. List( eminus, ana->Fill. List( piplus, ana->Fill. List( piminus, ana->Fill. List( gamma, "Electron. Tight. Plus", "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" ); "Electron. Tight. Minus", "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" ); "Pion. Loose. Plus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Pion. Loose. Minus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Neutral"); pi 0 ->Combine(gamma, gamma); jpsi->Combine(eplus, eminus, "J/psi"); pbarp->Combine(jpsi, piplus, piminus, pi 0, "pbarp. System"); if (ana->Mc. Truth. Match( pbarp[0] )) { Rho. Candidate *truth = pbarp[0]. Get. Mc. Truth(); . . . K. Götzen Glue. X PANDA Workshop 2019 9

Basic Analysis via ROOT Macro. . . Fair. Run. Ana *f. Run = new Fair. Run. Ana(); Object f. Run->Set. Input. File("pbarp_jpsi_pipipi 0. root"); for data access Pnd. Analysis *ana = new Pnd. Analysis(); Candidate Lists Rho. Cand. List eplus, eminus, piplus, piminus, gamma, pi 0, jpsi, pbarp; while ( ana->Get. Event() ) { ana->Fill. List( eplus, ana->Fill. List( eminus, ana->Fill. List( piplus, ana->Fill. List( piminus, ana->Fill. List( gamma, PID Selection & "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" Algorithms); "Electron. Tight. Plus", "Electron. Tight. Minus", "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" ); "Pion. Loose. Plus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Pion. Loose. Minus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Neutral"); pi 0 ->Combine(gamma, gamma); jpsi->Combine(eplus, eminus, "J/psi"); pbarp->Combine(jpsi, piplus, piminus, pi 0, "pbarp. System"); if (ana->Mc. Truth. Match( pbarp[0] )) { Rho. Candidate *truth = pbarp[0]. Get. Mc. Truth(); . . . K. Götzen Glue. X PANDA Workshop 2019 10

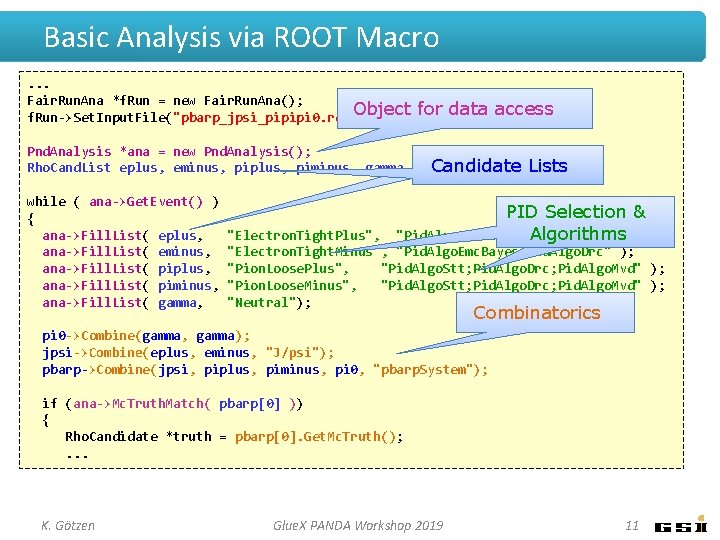
Basic Analysis via ROOT Macro. . . Fair. Run. Ana *f. Run = new Fair. Run. Ana(); Object f. Run->Set. Input. File("pbarp_jpsi_pipipi 0. root"); for data access Pnd. Analysis *ana = new Pnd. Analysis(); Candidate Lists Rho. Cand. List eplus, eminus, piplus, piminus, gamma, pi 0, jpsi, pbarp; while ( ana->Get. Event() ) { ana->Fill. List( eplus, ana->Fill. List( eminus, ana->Fill. List( piplus, ana->Fill. List( piminus, ana->Fill. List( gamma, PID Selection & "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" Algorithms); "Electron. Tight. Plus", "Electron. Tight. Minus", "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" ); "Pion. Loose. Plus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Pion. Loose. Minus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Neutral"); Combinatorics pi 0 ->Combine(gamma, gamma); jpsi->Combine(eplus, eminus, "J/psi"); pbarp->Combine(jpsi, piplus, piminus, pi 0, "pbarp. System"); if (ana->Mc. Truth. Match( pbarp[0] )) { Rho. Candidate *truth = pbarp[0]. Get. Mc. Truth(); . . . K. Götzen Glue. X PANDA Workshop 2019 11

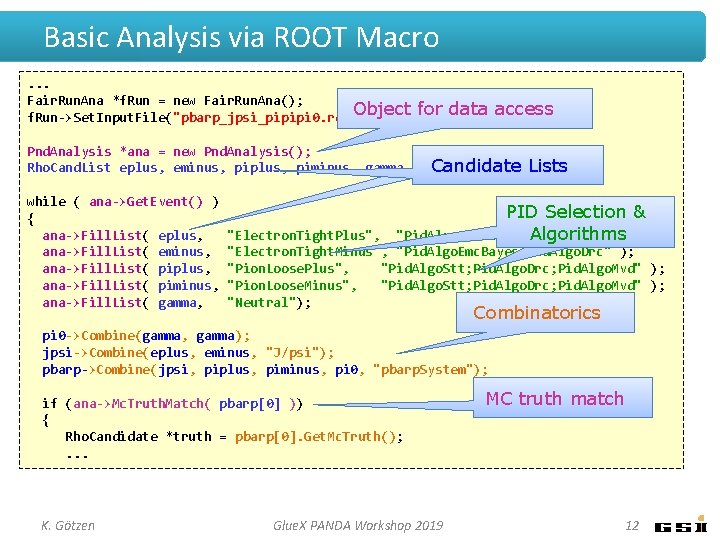
Basic Analysis via ROOT Macro. . . Fair. Run. Ana *f. Run = new Fair. Run. Ana(); Object f. Run->Set. Input. File("pbarp_jpsi_pipipi 0. root"); for data access Pnd. Analysis *ana = new Pnd. Analysis(); Candidate Lists Rho. Cand. List eplus, eminus, piplus, piminus, gamma, pi 0, jpsi, pbarp; while ( ana->Get. Event() ) { ana->Fill. List( eplus, ana->Fill. List( eminus, ana->Fill. List( piplus, ana->Fill. List( piminus, ana->Fill. List( gamma, PID Selection & "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" Algorithms); "Electron. Tight. Plus", "Electron. Tight. Minus", "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" ); "Pion. Loose. Plus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Pion. Loose. Minus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Neutral"); Combinatorics pi 0 ->Combine(gamma, gamma); jpsi->Combine(eplus, eminus, "J/psi"); pbarp->Combine(jpsi, piplus, piminus, pi 0, "pbarp. System"); if (ana->Mc. Truth. Match( pbarp[0] )) { Rho. Candidate *truth = pbarp[0]. Get. Mc. Truth(); . . . K. Götzen Glue. X PANDA Workshop 2019 MC truth match 12

Basic Analysis via ROOT Macro. . . Fair. Run. Ana *f. Run = new Fair. Run. Ana(); Object f. Run->Set. Input. File("pbarp_jpsi_pipipi 0. root"); for data access Pnd. Analysis *ana = new Pnd. Analysis(); Candidate Lists Rho. Cand. List eplus, eminus, piplus, piminus, gamma, pi 0, jpsi, pbarp; while ( ana->Get. Event() ) { ana->Fill. List( eplus, ana->Fill. List( eminus, ana->Fill. List( piplus, ana->Fill. List( piminus, ana->Fill. List( gamma, PID Selection & "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" Algorithms); "Electron. Tight. Plus", "Electron. Tight. Minus", "Pid. Algo. Emc. Bayes; Pid. Algo. Drc" ); "Pion. Loose. Plus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Pion. Loose. Minus", "Pid. Algo. Stt; Pid. Algo. Drc; Pid. Algo. Mvd" ); "Neutral"); n i s l i a t e D e r o M pi 0 ->Combine(gamma, B gamma); CKUP slides A jpsi->Combine(eplus, eminus, "J/psi"); Combinatorics pbarp->Combine(jpsi, piplus, piminus, pi 0, "pbarp. System"); if (ana->Mc. Truth. Match( pbarp[0] )) { Rho. Candidate *truth = pbarp[0]. Get. Mc. Truth(); . . . K. Götzen Glue. X PANDA Workshop 2019 MC truth match 13

Output and QA

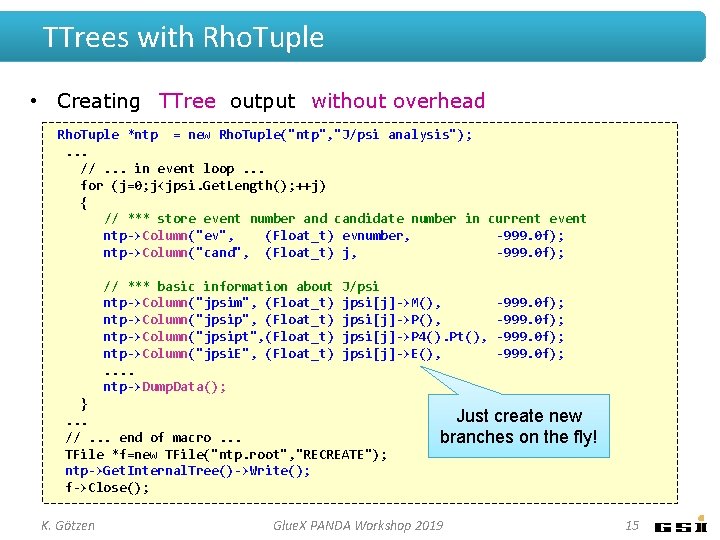
TTrees with Rho. Tuple • Creating TTree output without overhead Rho. Tuple *ntp = new Rho. Tuple("ntp", "J/psi analysis"); . . . //. . . in event loop. . . for (j=0; j<jpsi. Get. Length(); ++j) { // *** store event number and candidate number in current event ntp->Column("ev", (Float_t) evnumber, -999. 0 f); ntp->Column("cand", (Float_t) j, -999. 0 f); // *** basic information about ntp->Column("jpsim", (Float_t) ntp->Column("jpsipt", (Float_t) ntp->Column("jpsi. E", (Float_t). . ntp->Dump. Data(); J/psi jpsi[j]->M(), jpsi[j]->P 4(). Pt(), jpsi[j]->E(), }. . . //. . . end of macro. . . TFile *f=new TFile("ntp. root", "RECREATE"); ntp->Get. Internal. Tree()->Write(); f->Close(); K. Götzen -999. 0 f); Just create new branches on the fly! Glue. X PANDA Workshop 2019 15

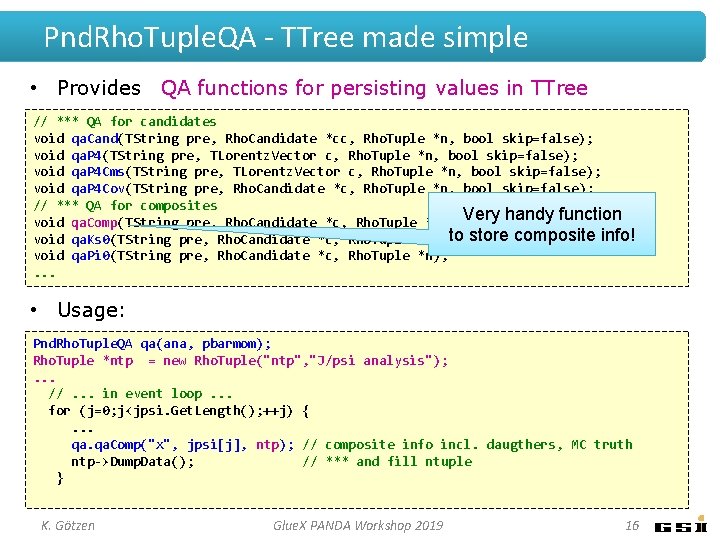
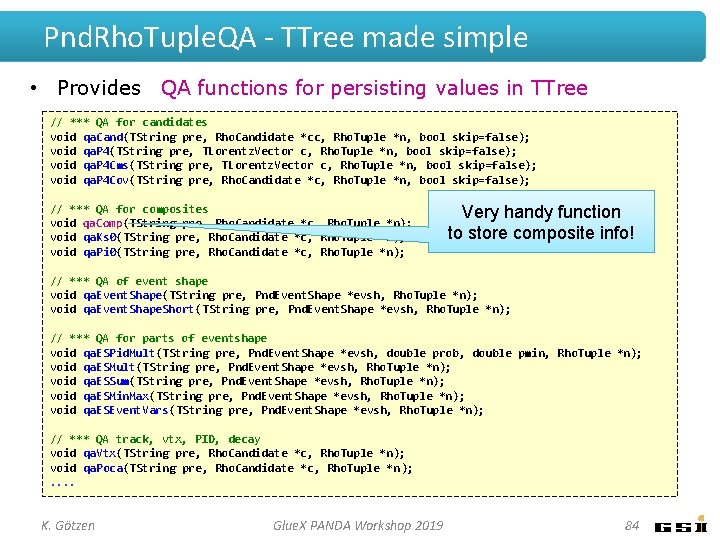
Pnd. Rho. Tuple. QA - TTree made simple • Provides QA functions for persisting values in TTree // *** QA for candidates void qa. Cand(TString pre, Rho. Candidate *cc, Rho. Tuple *n, bool skip=false); void qa. P 4(TString pre, TLorentz. Vector c, Rho. Tuple *n, bool skip=false); void qa. P 4 Cms(TString pre, TLorentz. Vector c, Rho. Tuple *n, bool skip=false); void qa. P 4 Cov(TString pre, Rho. Candidate *c, Rho. Tuple *n, bool skip=false); // *** QA for composites void qa. Comp(TString pre, Rho. Candidate *c, Rho. Tuple *n); Very handy function void qa. Ks 0(TString pre, Rho. Candidate *c, Rho. Tuple *n); to store composite info! void qa. Pi 0(TString pre, Rho. Candidate *c, Rho. Tuple *n); . . . • Usage: Pnd. Rho. Tuple. QA qa(ana, pbarmom); Rho. Tuple *ntp = new Rho. Tuple("ntp", "J/psi analysis"); . . . //. . . in event loop. . . for (j=0; j<jpsi. Get. Length(); ++j) {. . . qa. Comp("x", jpsi[j], ntp); // composite info incl. daugthers, MC truth ntp->Dump. Data(); // *** and fill ntuple } K. Götzen Glue. X PANDA Workshop 2019 16

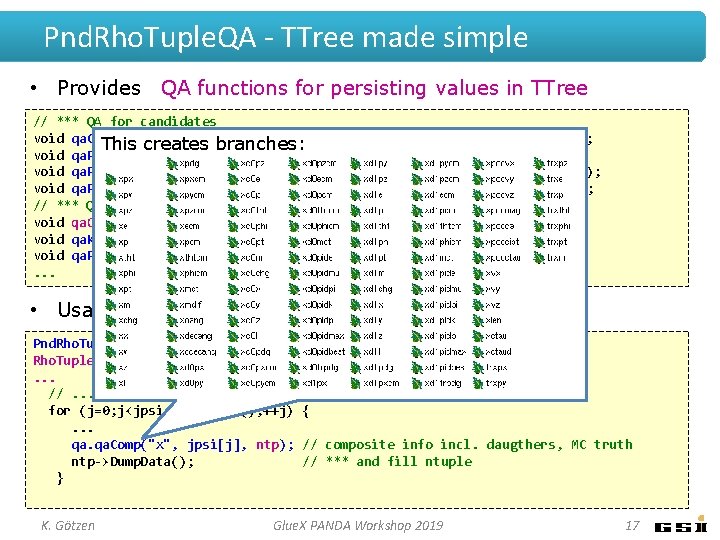
Pnd. Rho. Tuple. QA - TTree made simple • Provides QA functions for persisting values in TTree // *** QA for candidates void qa. Cand(TString pre, branches: Rho. Candidate *cc, Rho. Tuple *n, bool skip=false); This creates void qa. P 4(TString pre, TLorentz. Vector c, Rho. Tuple *n, bool skip=false); void qa. P 4 Cms(TString pre, TLorentz. Vector c, Rho. Tuple *n, bool skip=false); void qa. P 4 Cov(TString pre, Rho. Candidate *c, Rho. Tuple *n, bool skip=false); // *** QA for composites void qa. Comp(TString pre, Rho. Candidate *c, Rho. Tuple *n); void qa. Ks 0(TString pre, Rho. Candidate *c, Rho. Tuple *n); void qa. Pi 0(TString pre, Rho. Candidate *c, Rho. Tuple *n); . . . • Usage: Pnd. Rho. Tuple. QA qa(ana, pbarmom); Rho. Tuple *ntp = new Rho. Tuple("ntp", "J/psi analysis"); . . . //. . . in event loop. . . for (j=0; j<jpsi. Get. Length(); ++j) {. . . qa. Comp("x", jpsi[j], ntp); // composite info incl. daugthers, MC truth ntp->Dump. Data(); // *** and fill ntuple } K. Götzen Glue. X PANDA Workshop 2019 17

Pnd. Rho. Tuple. QA - TTree made simple • Provides QA functions for persisting values in TTree // *** QA for candidates void qa. Cand(TString pre, branches: Rho. Candidate *cc, Rho. Tuple *n, bool skip=false); This creates infos about daugther 0 void qa. P 4(TString pre, TLorentz. Vector c, Rho. Tuple *n, bool skip=false); void qa. P 4 Cms(TString pre, TLorentz. Vector c, Rho. Tuple *n, bool skip=false); void qa. P 4 Cov(TString pre, Rho. Candidate *c, Rho. Tuple *n, bool skip=false); // *** QA for composites void qa. Comp(TString pre, Rho. Candidate *c, Rho. Tuple *n); void qa. Ks 0(TString pre, Rho. Candidate *c, Rho. Tuple *n); void qa. Pi 0(TString pre, Rho. Candidate *c, Rho. Tuple *n); . . . • Usage: Pnd. Rho. Tuple. QA qa(ana, pbarmom); Rho. Tuple *ntp = new Rho. Tuple("ntp", "J/psi analysis"); . . . //. . . in event loop. . . for (j=0; j<jpsi. Get. Length(); ++j) {. . . qa. Comp("x", jpsi[j], ntp); // composite info incl. daugthers, MC truth ntp->Dump. Data(); // *** and fill ntuple } K. Götzen Glue. X PANDA Workshop 2019 18

Quick Analysis Tools

Quick Analysis • Based on Pnd. Simple. Combiner & Pnd. Simple. Combiner. Task – very simple and compact analysis approach – runs analysis in compiled Fair. Task (faster + more robust) -l -b -q 'quickana. C( • root Reco e. g. of mode p → J/ψ (→ μ+μ−) π+π− can be done by "pid_complete. root", 6. 232, "J/psi->mu+ mu-; pbarp. System->J/psi pi+ pi-", 0, "fit 4 c: fitvtx: mwin(J/psi)=0. 8" )' // // // input file p [Ge. V/c] decay tree reco #events (0=all) some parameters Attaching file pid_complete_ana. root as _file 0. . . root [1]. ls TFile** pid_complete_ana. root TFile* pid_complete_ana. root KEY: TFolder cbmout; 1 Main Output Folder KEY: TList Branch. List; 1 Doubly linked list KEY: Fair. File. Header; 1 KEY: TTree ntp 0; 1 J/psi->mu+ mu. KEY: TTree ntp 1; 1 pbarp. System->J/psi pi+ pi- K. Götzen Glue. X PANDA Workshop 2019 20

Quick Analysis • Based on Pnd. Simple. Combiner & Pnd. Simple. Combiner. Task – very simple and compact analysis approach – runs analysis in compiled Fair. Task (faster + more robust) Decay chain to be reco'd root -l -b -q 'quickana. C( + − • Reco e. g. of mode p → J/ψ (→ μ μ ) π π can be done by "pid_complete. root", 6. 232, "J/psi->mu+ mu-; pbarp. System->J/psi pi+ pi-", 0, "fit 4 c: fitvtx: mwin(J/psi)=0. 8" )' // // // input file p [Ge. V/c] decay tree reco #events (0=all) some parameters Reco options Attaching file pid_complete_ana. root as _file 0. . . root [1]. ls TFile** pid_complete_ana. root TFile* pid_complete_ana. root KEY: TFolder cbmout; 1 Main Output Folder KEY: TList Branch. List; 1 Doubly linked list KEY: Fair. File. Header; 1 KEY: TTree ntp 0; 1 J/psi->mu+ mu. KEY: TTree ntp 1; 1 pbarp. System->J/psi pi+ pi- K. Götzen Glue. X PANDA Workshop 2019 21

Quick Analysis • Based on Pnd. Simple. Combiner & Pnd. Simple. Combiner. Task – very simple and compact analysis approach – runs analysis in compiled Fair. Task (faster + more robust) -l -b -q 'quickana. C( • root Reco e. g. of mode p → J/ψ (→ μ+μ−) π+π− can be done by "pid_complete. root", 6. 232, "J/psi->mu+ mu-; pbarp. System->J/psi pi+ pi-", 0, "fit 4 c: fitvtx: mwin(J/psi)=0. 8" )' // // // input file p [Ge. V/c] decay tree reco #events (0=all) some parameters Attaching file pid_complete_ana. root as _file 0. . . root [1]. ls TFile** pid_complete_ana. root TFile* pid_complete_ana. root KEY: TFolder cbmout; 1 Main Output Folder KEY: TList Branch. List; 1 Doubly linked list KEY: Fair. File. Header; 1 KEY: TTree ntp 0; 1 J/psi->mu+ mu. KEY: TTree ntp 1; 1 pbarp. System->J/psi pi+ pi- K. Götzen Glue. X PANDA Workshop 2019 22

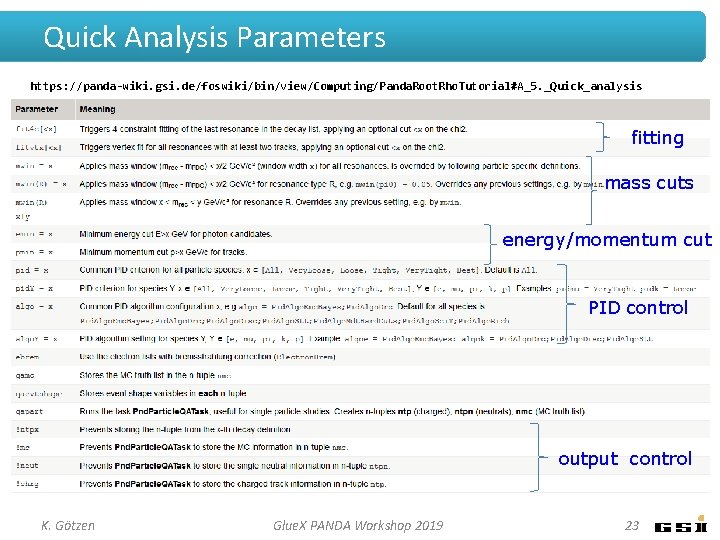
Quick Analysis Parameters https: //panda-wiki. gsi. de/foswiki/bin/view/Computing/Panda. Root. Rho. Tutorial#A_5. _Quick_analysis fitting mass cuts energy/momentum cut PID control output control K. Götzen Glue. X PANDA Workshop 2019 23

Quick Analysis Parameters https: //panda-wiki. gsi. de/foswiki/bin/view/Computing/Panda. Root. Rho. Tutorial#A_5. _Quick_analysis Our X(3872) scan analysis was completely done with quickana. C. . . and is published now! [EPJ A 55 (2019) 42] K. Götzen Glue. X PANDA Workshop 2019 24

Fast Simulation

Fast Simulations • Idea: Particle smearing instead Geant simulation • Simulate large number of events time in short • Very simple configuration • Study of – Reaction kinematics – Detector configurations • Output analysed with same analysis code as for full simulation K. Götzen Glue. X PANDA Workshop 2019 26

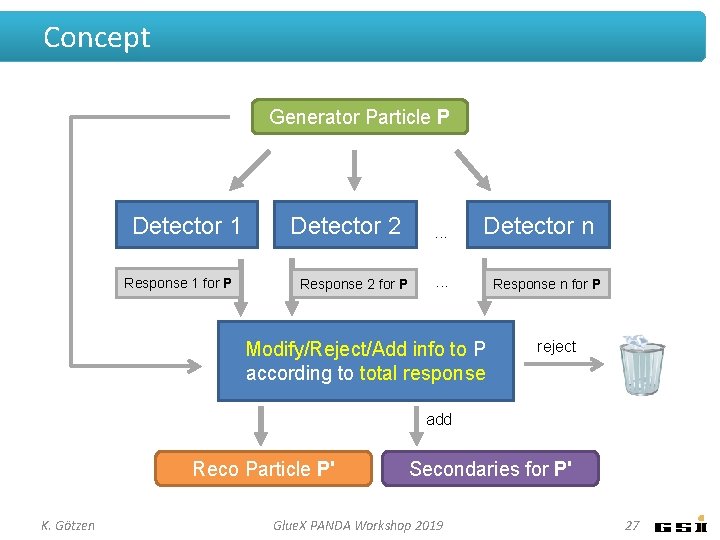
Concept Generator Particle P Detector 1 Response 1 for P Detector 2 Response 2 for P . . . Detector n . . . Modify/Reject/Add info to P according to total response Response n for P reject add Reco Particle P' K. Götzen Secondaries for P' Glue. X PANDA Workshop 2019 27

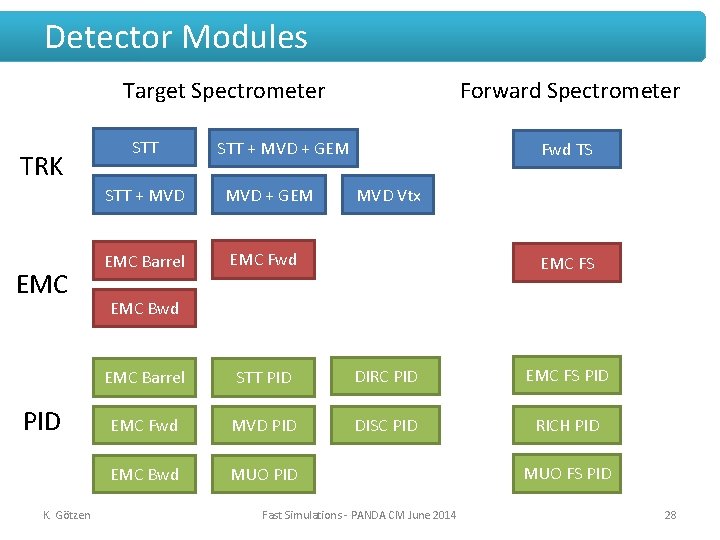
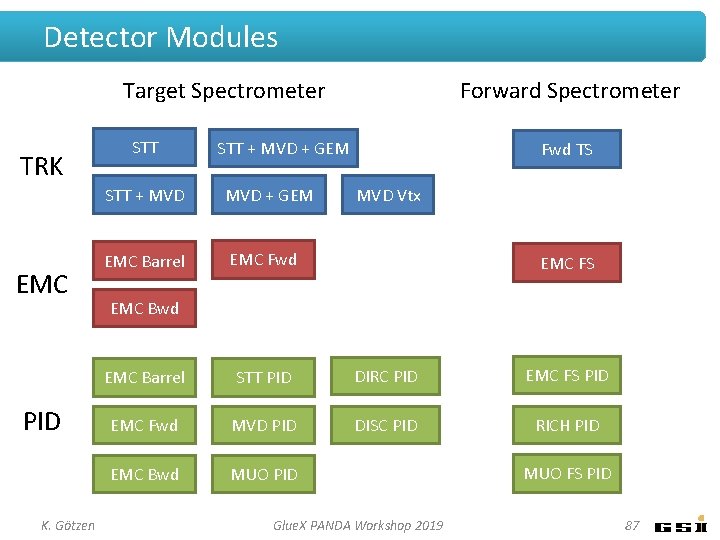
Detector Modules Target Spectrometer TRK EMC PID K. Götzen STT Forward Spectrometer STT + MVD + GEM EMC Barrel EMC Fwd TS MVD Vtx EMC FS EMC Bwd EMC Barrel STT PID DIRC PID EMC FS PID EMC Fwd MVD PID DISC PID RICH PID EMC Bwd MUO PID Fast Simulations - PANDA CM June 2014 MUO FS PID 28

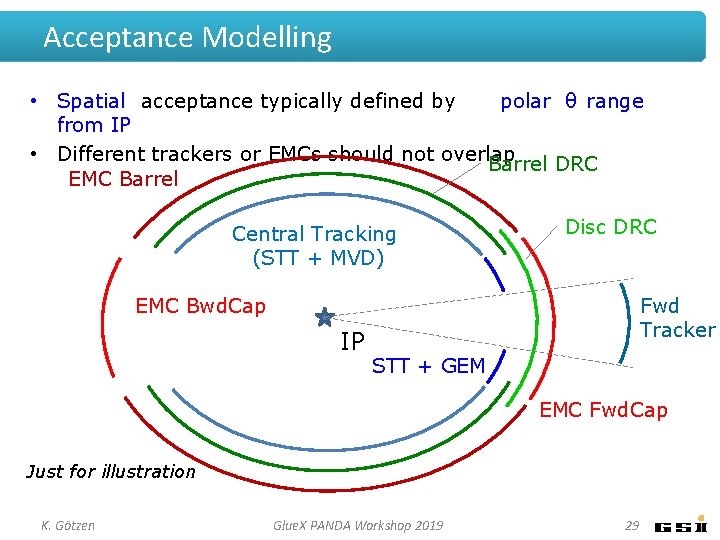
Acceptance Modelling • Spatial acceptance typically defined by polar θ range from IP • Different trackers or EMCs should not overlap Barrel DRC EMC Barrel Central Tracking (STT + MVD) Disc DRC EMC Bwd. Cap IP Fwd Tracker STT + GEM EMC Fwd. Cap Just for illustration K. Götzen Glue. X PANDA Workshop 2019 29

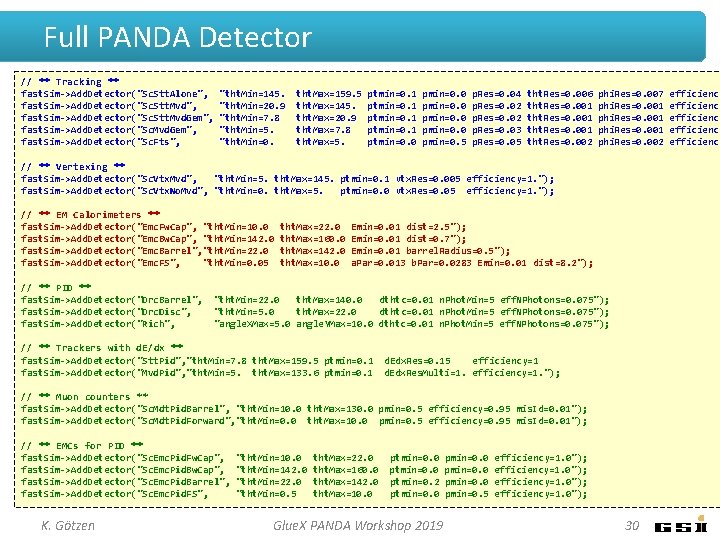
Full PANDA Detector // ** Tracking ** fast. Sim->Add. Detector("Sc. Stt. Alone", fast. Sim->Add. Detector("Sc. Stt. Mvd. Gem", fast. Sim->Add. Detector("Sc. Fts", "tht. Min=145. "tht. Min=20. 9 "tht. Min=7. 8 "tht. Min=5. "tht. Min=0. tht. Max=159. 5 tht. Max=145. tht. Max=20. 9 tht. Max=7. 8 tht. Max=5. ptmin=0. 1 ptmin=0. 0 pmin=0. 5 p. Res=0. 04 p. Res=0. 02 p. Res=0. 03 p. Res=0. 05 tht. Res=0. 006 tht. Res=0. 001 tht. Res=0. 002 phi. Res=0. 007 phi. Res=0. 001 phi. Res=0. 002 // ** Vertexing ** fast. Sim->Add. Detector("Sc. Vtx. Mvd", "tht. Min=5. tht. Max=145. ptmin=0. 1 vtx. Res=0. 005 efficiency=1. "); fast. Sim->Add. Detector("Sc. Vtx. No. Mvd", "tht. Min=0. tht. Max=5. ptmin=0. 0 vtx. Res=0. 05 efficiency=1. "); // ** EM Calorimeters ** fast. Sim->Add. Detector("Emc. Fw. Cap", "tht. Min=10. 0 fast. Sim->Add. Detector("Emc. Bw. Cap", "tht. Min=142. 0 fast. Sim->Add. Detector("Emc. Barrel", "tht. Min=22. 0 fast. Sim->Add. Detector("Emc. FS", "tht. Min=0. 05 // ** PID ** fast. Sim->Add. Detector("Drc. Barrel", fast. Sim->Add. Detector("Drc. Disc", fast. Sim->Add. Detector("Rich", tht. Max=22. 0 tht. Max=160. 0 tht. Max=142. 0 tht. Max=10. 0 Emin=0. 01 dist=2. 5"); Emin=0. 01 dist=0. 7"); Emin=0. 01 barrel. Radius=0. 5"); a. Par=0. 013 b. Par=0. 0283 Emin=0. 01 dist=8. 2"); "tht. Min=22. 0 tht. Max=140. 0 dthtc=0. 01 n. Phot. Min=5 eff. NPhotons=0. 075"); "tht. Min=5. 0 tht. Max=22. 0 dthtc=0. 01 n. Phot. Min=5 eff. NPhotons=0. 075"); "angle. XMax=5. 0 angle. YMax=10. 0 dthtc=0. 01 n. Phot. Min=5 eff. NPhotons=0. 075"); // ** Trackers with d. E/dx ** fast. Sim->Add. Detector("Stt. Pid", "tht. Min=7. 8 tht. Max=159. 5 ptmin=0. 1 fast. Sim->Add. Detector("Mvd. Pid", "tht. Min=5. tht. Max=133. 6 ptmin=0. 1 d. Edx. Res=0. 15 efficiency=1 d. Edx. Res. Multi=1. efficiency=1. "); // ** Muon counters ** fast. Sim->Add. Detector("Sc. Mdt. Pid. Barrel", "tht. Min=10. 0 tht. Max=130. 0 pmin=0. 5 efficiency=0. 95 mis. Id=0. 01"); fast. Sim->Add. Detector("Sc. Mdt. Pid. Forward", "tht. Min=0. 0 tht. Max=10. 0 pmin=0. 5 efficiency=0. 95 mis. Id=0. 01"); // ** EMCs for PID ** fast. Sim->Add. Detector("Sc. Emc. Pid. Fw. Cap", fast. Sim->Add. Detector("Sc. Emc. Pid. Barrel", fast. Sim->Add. Detector("Sc. Emc. Pid. FS", K. Götzen "tht. Min=10. 0 "tht. Min=142. 0 "tht. Min=22. 0 "tht. Min=0. 5 tht. Max=22. 0 tht. Max=160. 0 tht. Max=142. 0 tht. Max=10. 0 ptmin=0. 2 ptmin=0. 0 Glue. X PANDA Workshop 2019 pmin=0. 0 pmin=0. 5 efficiency=1. 0"); 30 efficiency efficiency

Full PANDA Detector // ** Tracking ** fast. Sim->Add. Detector("Sc. Stt. Alone", fast. Sim->Add. Detector("Sc. Stt. Mvd. Gem", fast. Sim->Add. Detector("Sc. Fts", "tht. Min=145. "tht. Min=20. 9 "tht. Min=7. 8 "tht. Min=5. "tht. Min=0. tht. Max=159. 5 tht. Max=145. tht. Max=20. 9 tht. Max=7. 8 tht. Max=5. ptmin=0. 1 ptmin=0. 0 pmin=0. 5 p. Res=0. 04 p. Res=0. 02 p. Res=0. 03 p. Res=0. 05 tht. Res=0. 006 tht. Res=0. 001 tht. Res=0. 002 phi. Res=0. 007 phi. Res=0. 001 phi. Res=0. 002 // ** Vertexing ** fast. Sim->Add. Detector("Sc. Vtx. Mvd", "tht. Min=5. tht. Max=145. ptmin=0. 1 vtx. Res=0. 005 efficiency=1. "); fast. Sim->Add. Detector("Sc. Vtx. No. Mvd", "tht. Min=0. tht. Max=5. ptmin=0. 0 vtx. Res=0. 05 efficiency=1. "); // ** EM Calorimeters ** fast. Sim->Add. Detector("Emc. Fw. Cap", "tht. Min=10. 0 fast. Sim->Add. Detector("Emc. Bw. Cap", "tht. Min=142. 0 fast. Sim->Add. Detector("Emc. Barrel", "tht. Min=22. 0 fast. Sim->Add. Detector("Emc. FS", "tht. Min=0. 05 tht. Max=22. 0 tht. Max=160. 0 tht. Max=142. 0 tht. Max=10. 0 Emin=0. 01 dist=2. 5"); Emin=0. 01 dist=0. 7"); Emin=0. 01 barrel. Radius=0. 5"); a. Par=0. 013 b. Par=0. 0283 Emin=0. 01 dist=8. 2"); Glue. X p u t e s d l u o c Probably e : -) m i t t r o h s n i Detector // ** PID ** fast. Sim->Add. Detector("Drc. Barrel", fast. Sim->Add. Detector("Drc. Disc", fast. Sim->Add. Detector("Rich", "tht. Min=22. 0 tht. Max=140. 0 dthtc=0. 01 n. Phot. Min=5 eff. NPhotons=0. 075"); "tht. Min=5. 0 tht. Max=22. 0 dthtc=0. 01 n. Phot. Min=5 eff. NPhotons=0. 075"); "angle. XMax=5. 0 angle. YMax=10. 0 dthtc=0. 01 n. Phot. Min=5 eff. NPhotons=0. 075"); // ** Trackers with d. E/dx ** fast. Sim->Add. Detector("Stt. Pid", "tht. Min=7. 8 tht. Max=159. 5 ptmin=0. 1 d. Edx. Res=0. 15 efficiency=1 fast. Sim->Add. Detector("Mvd. Pid", "tht. Min=5. tht. Max=133. 6 ptmin=0. 1 d. Edx. Res. Multi=1. efficiency=1. "); // ** Muon counters ** fast. Sim->Add. Detector("Sc. Mdt. Pid. Barrel", "tht. Min=10. 0 tht. Max=130. 0 pmin=0. 5 efficiency=0. 95 mis. Id=0. 01"); fast. Sim->Add. Detector("Sc. Mdt. Pid. Forward", "tht. Min=0. 0 tht. Max=10. 0 pmin=0. 5 efficiency=0. 95 mis. Id=0. 01"); // ** EMCs for PID ** fast. Sim->Add. Detector("Sc. Emc. Pid. Fw. Cap", fast. Sim->Add. Detector("Sc. Emc. Pid. Barrel", fast. Sim->Add. Detector("Sc. Emc. Pid. FS", K. Götzen "tht. Min=10. 0 "tht. Min=142. 0 "tht. Min=22. 0 "tht. Min=0. 5 tht. Max=22. 0 ptmin=0. 0 pmin=0. 0 efficiency=1. 0"); tht. Max=160. 0 ptmin=0. 0 pmin=0. 0 efficiency=1. 0"); tht. Max=142. 0 ptmin=0. 2 pmin=0. 0 efficiency=1. 0"); tht. Max=10. 0 ptmin=0. 0 pmin=0. 5 efficiency=1. 0"); Glue. X PANDA Workshop 2019 31 efficiency efficiency

Detector Parameters/Response • Detector parameters – Global detection efficiency ε – Spatial acceptance in θ (sometimes φ ) – Kinematic acceptance ( p min , p t, min , E min , . . . ) – Resolutions: dp/p, d. E/E, dt/t, d θ, dφ , vertex – Detector specific parameters ( geometry, # layers, granularity , . . . ) • Detector responses – Accept/reject particle – Current resolution dp/p, d. E/E, dt/t, d θ, dφ , vertex – PID probabilities ( P e , P μ , P π , P K , P p ) – Raw PID information ( E EMC , θ C, d. E/dx, L iron , . . . ) • Total response – created from individual responses + applied to particle K. Götzen Glue. X PANDA Workshop 2019 32

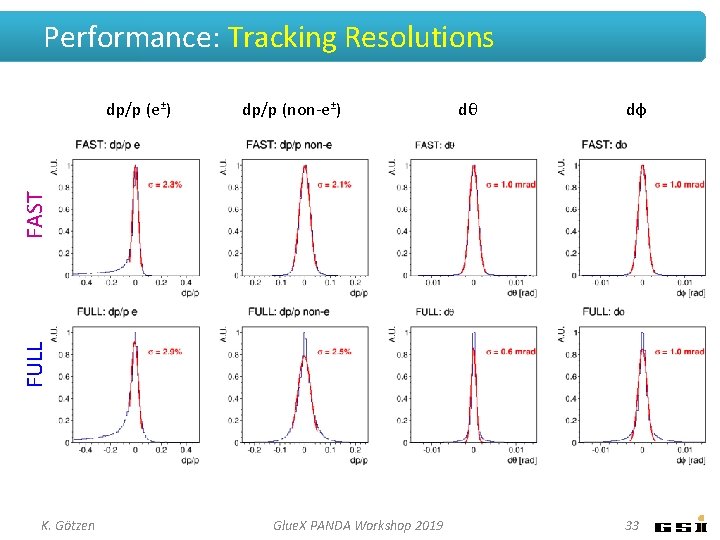
Performance: Tracking Resolutions dp/p (non-e±) dθ dφ FULL FAST dp/p (e±) K. Götzen Glue. X PANDA Workshop 2019 33

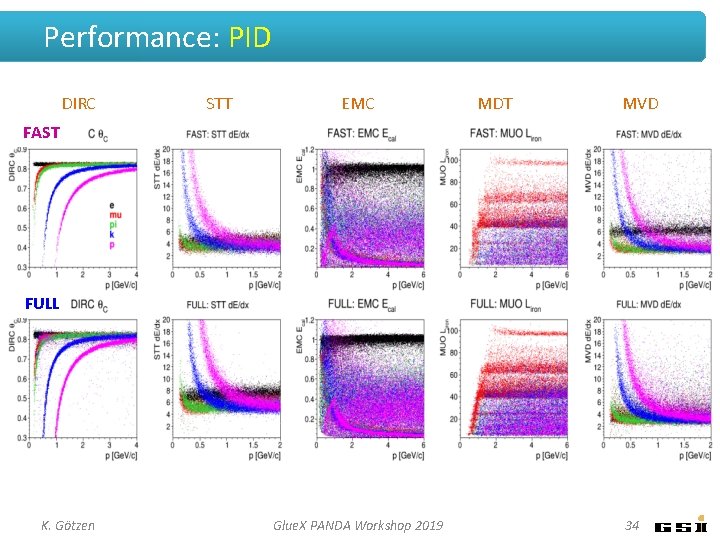
Performance: PID DIRC STT EMC MDT MVD FAST FULL K. Götzen Glue. X PANDA Workshop 2019 34

Combining Fast Simulation with Quick Analysis

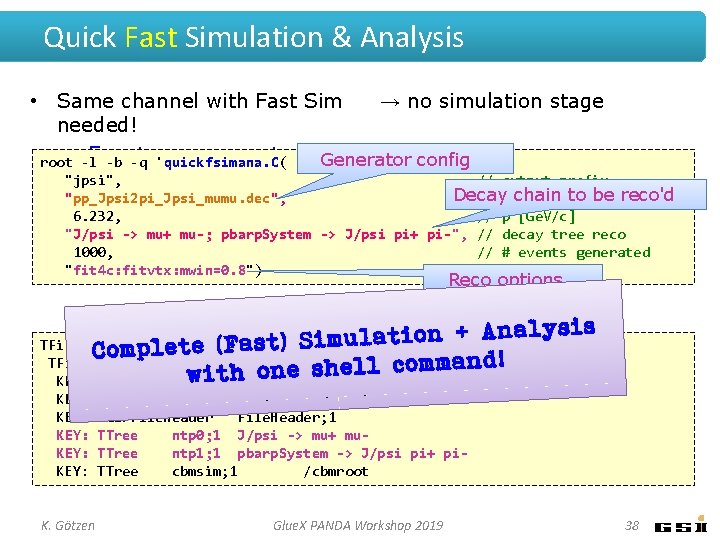
Quick Fast Simulation & Analysis • Same channel with Fast Sim → no simulation stage needed! Events are generated on-the-fly root⇒-l -b -q 'quickfsimana. C( "jpsi", // output prefix "pp_Jpsi 2 pi_Jpsi_mumu. dec", // decay file 6. 232, // p [Ge. V/c] "J/psi -> mu+ mu-; pbarp. System -> J/psi pi+ pi-", // decay tree reco 1000, // # events generated "fit 4 c: fitvtx: mwin=0. 8") // parameters TFile** jpsi_0_ana. root TFile* jpsi_0_ana. root KEY: TFolder cbmroot; 1 Main Folder KEY: TList Branch. List; 1 Doubly linked list KEY: Fair. File. Header; 1 KEY: TTree ntp 0; 1 J/psi -> mu+ mu. KEY: TTree ntp 1; 1 pbarp. System -> J/psi pi+ pi. KEY: TTree cbmsim; 1 /cbmroot K. Götzen Glue. X PANDA Workshop 2019 36

Quick Fast Simulation & Analysis • Same channel with Fast Sim → no simulation stage needed! Events are generated on-the-fly Generator config root⇒-l -b -q 'quickfsimana. C( "jpsi", // output prefix Decay chain to be reco'd "pp_Jpsi 2 pi_Jpsi_mumu. dec", // decay file 6. 232, // p [Ge. V/c] "J/psi -> mu+ mu-; pbarp. System -> J/psi pi+ pi-", // decay tree reco 1000, // # events generated "fit 4 c: fitvtx: mwin=0. 8") // parameters Reco options TFile** jpsi_0_ana. root TFile* jpsi_0_ana. root KEY: TFolder cbmroot; 1 Main Folder KEY: TList Branch. List; 1 Doubly linked list KEY: Fair. File. Header; 1 KEY: TTree ntp 0; 1 J/psi -> mu+ mu. KEY: TTree ntp 1; 1 pbarp. System -> J/psi pi+ pi. KEY: TTree cbmsim; 1 /cbmroot K. Götzen Glue. X PANDA Workshop 2019 37

Quick Fast Simulation & Analysis • Same channel with Fast Sim → no simulation stage needed! Events are generated on-the-fly Generator config root⇒-l -b -q 'quickfsimana. C( "jpsi", // output prefix Decay chain to be reco'd "pp_Jpsi 2 pi_Jpsi_mumu. dec", // decay file 6. 232, // p [Ge. V/c] "J/psi -> mu+ mu-; pbarp. System -> J/psi pi+ pi-", // decay tree reco 1000, // # events generated "fit 4 c: fitvtx: mwin=0. 8") // parameters Reco options lysis a n A + n o i t a l u m i S TFile** omplejpsi_0_ana. root te (Fast) C d! TFile* jpsi_0_ana. root n a m m o c l l e h s e n o with KEY: TFolder cbmroot; 1 Main Folder KEY: KEY: K. Götzen TList Branch. List; 1 Doubly linked list Fair. File. Header; 1 TTree ntp 0; 1 J/psi -> mu+ mu. TTree ntp 1; 1 pbarp. System -> J/psi pi+ pi. TTree cbmsim; 1 /cbmroot Glue. X PANDA Workshop 2019 38

Event Generators Stand-alone Usage

Important Particle Generators in PANDA • Evt. Gen (Pnd. Evt. Gen. Direct / simple. Evt. Gen ) – Generate signal reactions or specific backgrounds • DPM - Dual Parton Model ( Pnd. Dpm. Direct – Study p background reactions / DPMGen ) • FTF (Pnd. Ftf. Direct / FTFGen ) – Study p and A background reactions • Particle Gun (Pnd. Box. Generator ) – Single tracks for acceptance, efficiency, resolution studies • Many others not discussed here – Gi. Buu, Ur. Qmd, Fluka, Pythia, . . . K. Götzen PANDA Computing Workshop - Thailand 40

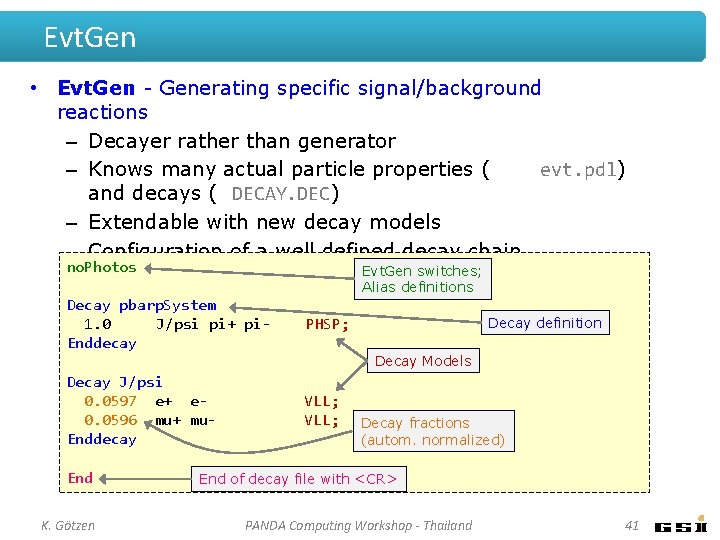
Evt. Gen • Evt. Gen - Generating specific signal/background reactions – Decayer rather than generator – Knows many actual particle properties ( evt. pdl) and decays ( DECAY. DEC) – Extendable with new decay models – Configuration of a well defined decay chain no. Photos Evt. Gen switches; Alias definitions Decay pbarp. System 1. 0 J/psi pi+ pi. Enddecay PHSP; Decay J/psi 0. 0597 e+ e 0. 0596 mu+ mu. Enddecay VLL; End K. Götzen Decay definition Decay Models Decay fractions (autom. normalized) End of decay file with <CR> PANDA Computing Workshop - Thailand 41

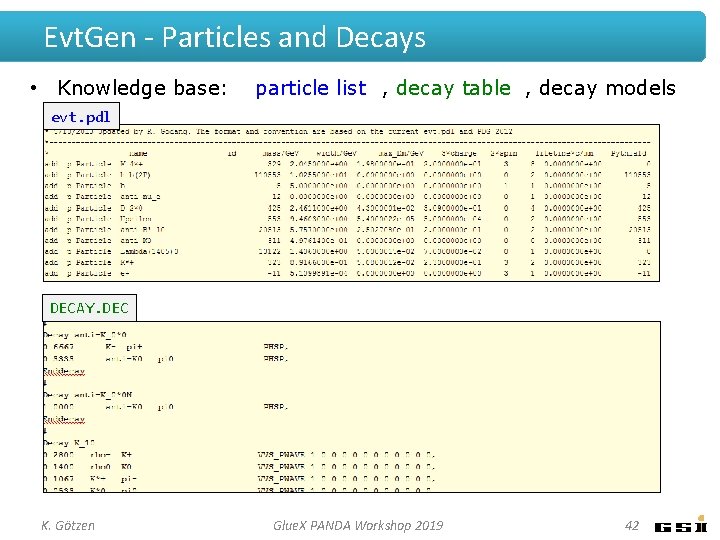
Evt. Gen - Particles and Decays • Knowledge base: particle list , decay table , decay models evt. pdl DECAY. DEC K. Götzen Glue. X PANDA Workshop 2019 42

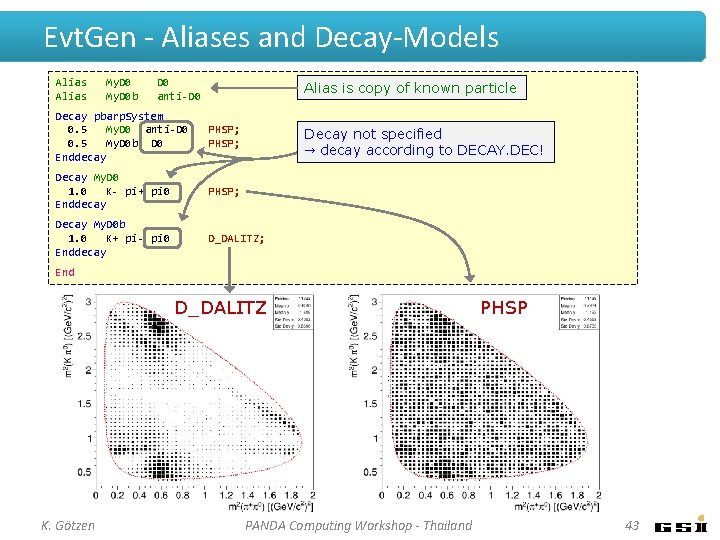
Evt. Gen - Aliases and Decay-Models Alias My. D 0 b D 0 anti-D 0 Alias is copy of known particle Decay pbarp. System 0. 5 My. D 0 anti-D 0 0. 5 My. D 0 b D 0 Enddecay PHSP; Decay My. D 0 1. 0 K- pi+ pi 0 Enddecay PHSP; Decay My. D 0 b 1. 0 K+ pi- pi 0 Enddecay D_DALITZ; Decay not specified → decay according to DECAY. DEC! End D_DALITZ K. Götzen PANDA Computing Workshop - Thailand PHSP 43

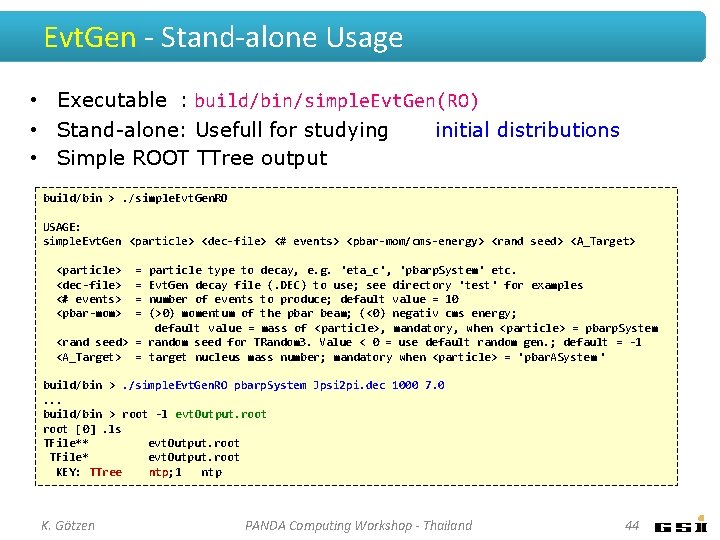
Evt. Gen - Stand-alone Usage • Executable : build/bin/simple. Evt. Gen(RO) • Stand-alone: Usefull for studying initial distributions • Simple ROOT TTree output build/bin >. /simple. Evt. Gen. RO USAGE: simple. Evt. Gen <particle> <dec-file> <# events> <pbar-mom/cms-energy> <rand seed> <A_Target> <particle> <dec-file> <# events> <pbar-mom> = = particle type to decay, e. g. 'eta_c', 'pbarp. System' etc. Evt. Gen decay file (. DEC) to use; see directory 'test' for examples number of events to produce; default value = 10 (>0) momentum of the pbar beam; (<0) negativ cms energy; default value = mass of <particle>, mandatory, when <particle> = pbarp. System <rand seed> = random seed for TRandom 3. Value < 0 = use default random gen. ; default = -1 <A_Target> = target nucleus mass number; mandatory when <particle> = 'pbar. ASystem ' build/bin >. /simple. Evt. Gen. RO pbarp. System Jpsi 2 pi. dec 1000 7. 0. . . build/bin > root -l evt. Output. root [0]. ls TFile** evt. Output. root TFile* evt. Output. root KEY: TTree ntp; 1 ntp K. Götzen PANDA Computing Workshop - Thailand 44

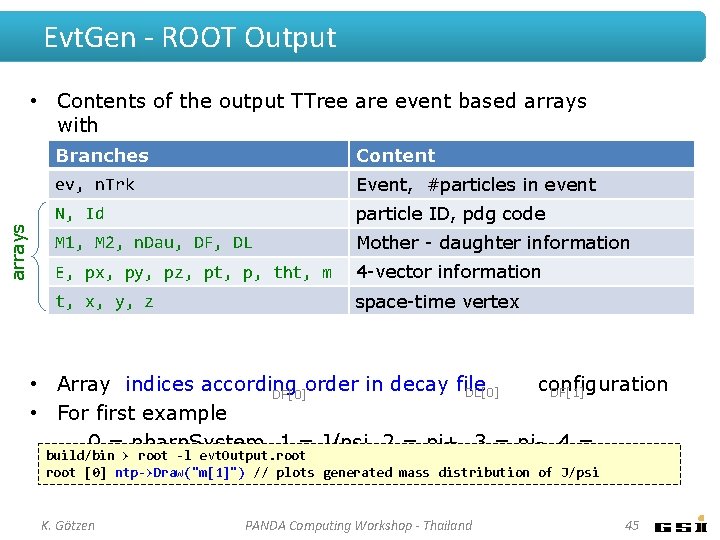
Evt. Gen - ROOT Output arrays • Contents of the output TTree are event based arrays with Branches Content ev, n. Trk Event, #particles in event N, Id particle ID, pdg code M 1, M 2, n. Dau, DF, DL Mother - daughter information E, px, py, pz, pt, p, tht, m 4 -vector information t, x, y, z space-time vertex • Array indices according order in decay file configuration DL[0] DF[1] DF[0] • For first example – 0 = pbarp. System, 1 = J/psi, 2 = pi+, 3 = pi-, 4 = build/bin > root -l evt. Output. root mu+, . . . root [0] ntp->Draw("m[1]") // plots generated mass distribution of J/psi K. Götzen PANDA Computing Workshop - Thailand 45

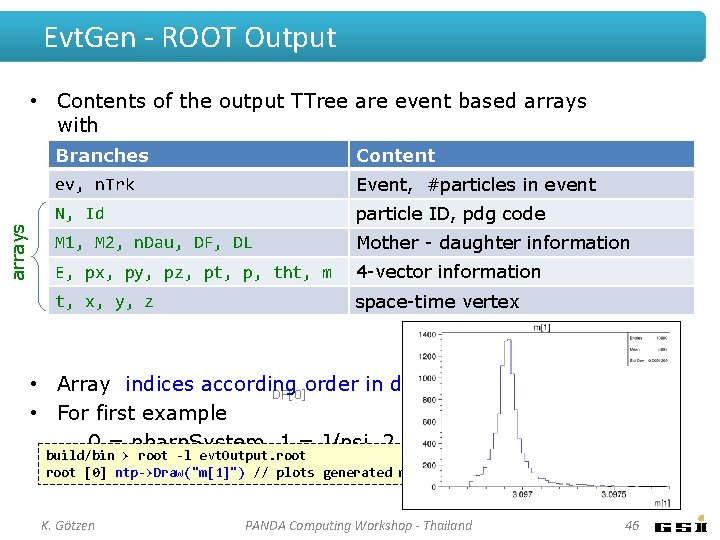
Evt. Gen - ROOT Output arrays • Contents of the output TTree are event based arrays with Branches Content ev, n. Trk Event, #particles in event N, Id particle ID, pdg code M 1, M 2, n. Dau, DF, DL Mother - daughter information E, px, py, pz, pt, p, tht, m 4 -vector information t, x, y, z space-time vertex • Array indices according order in decay file configuration DL[0] DF[1] DF[0] • For first example – 0 = pbarp. System, 1 = J/psi, 2 = pi+, 3 = pi-, 4 = build/bin > root -l evt. Output. root mu+, . . . root [0] ntp->Draw("m[1]") // plots generated mass distribution of J/psi K. Götzen PANDA Computing Workshop - Thailand 46

DPM - Stand-alone Usage • Stand-alone executable: build/bin/DPMGen • I nput parameters: – Random seed – Beam momentum (for PANDA: 1. 5. . . 15 Ge. V/c) – M ode (0 = inelastic, 1 = inel. + elastic, 2 = elastic) – Un/stable settings (e. g. '111 0' lets π0 be decayed by DPM) – Number of events • Output: – ROOT file containing K. Götzen TTree 'data' with PANDA Computing Workshop - Thailand TParticles 47

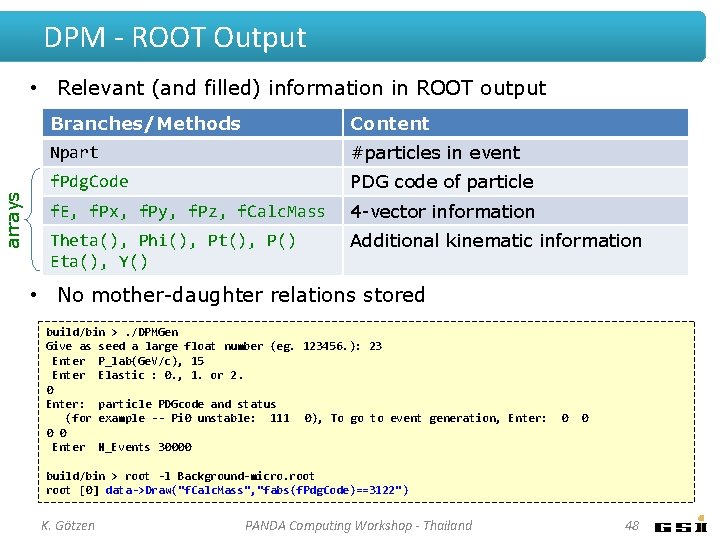
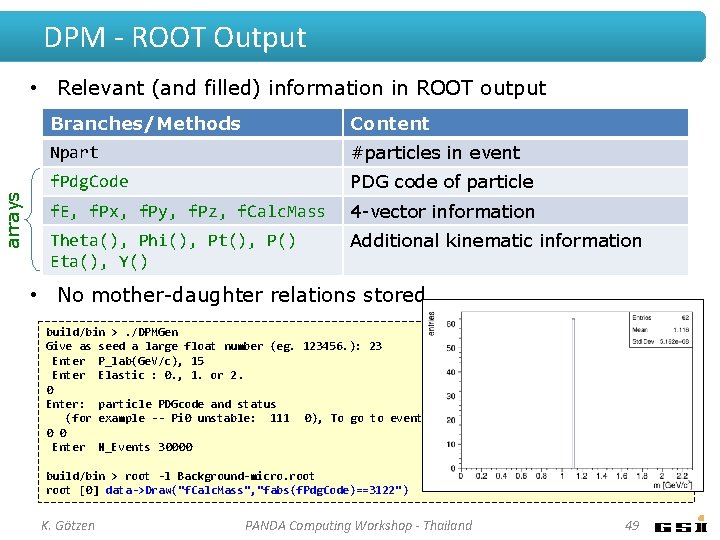
DPM - ROOT Output arrays • Relevant (and filled) information in ROOT output Branches/Methods Content Npart #particles in event f. Pdg. Code PDG code of particle f. E, f. Px, f. Py, f. Pz, f. Calc. Mass 4 -vector information Theta(), Phi(), Pt(), P() Eta(), Y() Additional kinematic information • No mother-daughter relations stored build/bin >. /DPMGen Give as seed a large float number (eg. 123456. ): 23 Enter P_lab(Ge. V/c), 15 Enter Elastic : 0. , 1. or 2. 0 Enter: particle PDGcode and status (for example -- Pi 0 unstable: 111 0), To go to event generation, Enter: 0 0 Enter N_Events 30000 0 0 build/bin > root -l Background-micro. root [0] data->Draw("f. Calc. Mass", "fabs(f. Pdg. Code)==3122") K. Götzen PANDA Computing Workshop - Thailand 48

DPM - ROOT Output arrays • Relevant (and filled) information in ROOT output Branches/Methods Content Npart #particles in event f. Pdg. Code PDG code of particle f. E, f. Px, f. Py, f. Pz, f. Calc. Mass 4 -vector information Theta(), Phi(), Pt(), P() Eta(), Y() Additional kinematic information • No mother-daughter relations stored build/bin >. /DPMGen Give as seed a large float number (eg. 123456. ): 23 Enter P_lab(Ge. V/c), 15 Enter Elastic : 0. , 1. or 2. 0 Enter: particle PDGcode and status (for example -- Pi 0 unstable: 111 0), To go to event generation, Enter: 0 0 Enter N_Events 30000 0 0 build/bin > root -l Background-micro. root [0] data->Draw("f. Calc. Mass", "fabs(f. Pdg. Code)==3122") K. Götzen PANDA Computing Workshop - Thailand 49

Event Filtering

Event Filtering • K. Götzen PANDA Computing Workshop - Thailand 51

Event Filtering in Panda. Root Pnd. Filtered. Primary. Generator (w/ Pnd. Smp. Filt, Pnd. Smp. Cand) • Complete Normal Form (DNF = logic via , Disjunctive e. g. ) • Filter on: Multiplicity, kinematic quantities, composite mass • Filterset: Individual filters combined with ' &&' (AND) • Multiple filtersets combined with '||' (OR) • Negate individual filters with '!' (NOT) • Example: Pnd. Filtered. Primary. Generator* prim. Gen Pnd. Filtered. Primary. Generator(); !N trk ≤ 3 AND N gam ≤ 6 AND= new N(2. 0<M(e+ e-)<4. 0 Ge. V/c 2 ) ≥ 1 prim. Gen->Add. Filter("!(t+; . . 3) && (gam ; . . 6) && M(e+ e- ; m[3. 0, 2. 0])"); Aug. 14, 2018 K. Götzen - Panda. Root Meeting (No number range given defaults to '1. . ' = at least one) 52

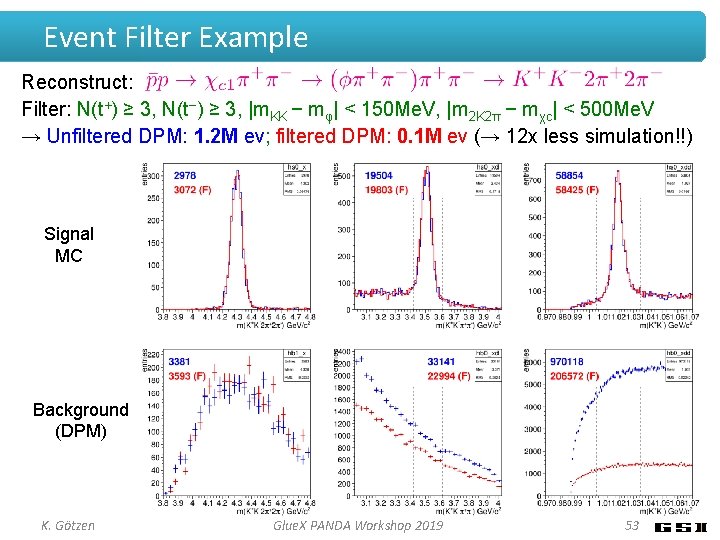
Event Filter Example Reconstruct: Filter: N(t+) ≥ 3, N(t−) ≥ 3, |m. KK − mφ| < 150 Me. V, |m 2 K 2π − mχc| < 500 Me. V → Unfiltered DPM: 1. 2 M ev; filtered DPM: 0. 1 M ev (→ 12 x less simulation!!) Signal MC Background (DPM) K. Götzen Glue. X PANDA Workshop 2019 53

Glue. X DSelector Flat. Tree Extension

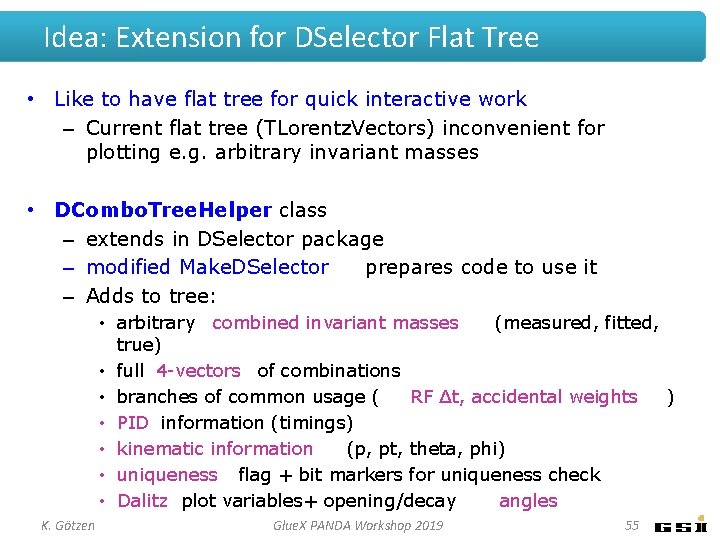
Idea: Extension for DSelector Flat Tree • Like to have flat tree for quick interactive work – Current flat tree (TLorentz. Vectors) inconvenient for plotting e. g. arbitrary invariant masses • DCombo. Tree. Helper class – extends in DSelector package – modified Make. DSelector prepares code to use it – Adds to tree: • arbitrary combined invariant masses (measured, fitted, true) • full 4 -vectors of combinations • branches of common usage ( RF ∆t, accidental weights ) • PID information (timings) • kinematic information (p, pt, theta, phi) • uniqueness flag + bit markers for uniqueness check • Dalitz plot variables+ opening/decay angles K. Götzen Glue. X PANDA Workshop 2019 55

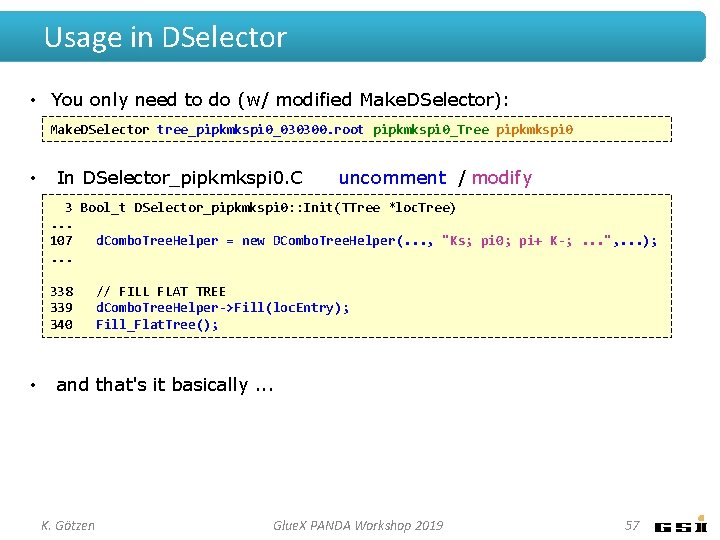
Usage in DSelector • Inside DSelector_ xyz : : Init() [here e. g. for input: pipkmkspi 0] . . . //CREATE HELPER AND INITIALIZE WITH DESIRED COMBINATIONS TO BE STORED d. Combo. Tree. Helper = new DCombo. Tree. Helper( d. Tree. Interface, d. Combo. Wrapper, d. Flat. Tree. Interface, // the interfaces "Ks; pi 0; pi+ K-; Ks pi+; Ks K-; p Ks; p pi 0", // combinations to be stored d. Thrown. Wrapper, // interface MC match "p 4: acc: pid: kin: marker: angle: dalitz"); // options • Adds branches (based on options): – always charge – p 4 – acc – pid – kin – marker – angle – dalitz K. Götzen : Inv. masses (_m, _mf, _mt), MM² (mm 2), unique flag : : : : (_uni, _unifs), 4 -vectors of combinations as TLorentz. Vector (_meas, _kin, _thr) RF dt, weights for 2, 4, and 8 acc. bunches (rf_dt, w 2, w 4, w 8) dt TOF , dt BCAL , dt FCAL (_dttof, _dtbcal, _dtfcal) kinematics of combinations (_p, _pt, _tht, _phi, beam_e, beam_z) bit marker for used final states (idxs_) opening/decay angles (_dec, _cdec, _oang) Dalitz vars (_m 01, _m 02, _m 12, _dal 01, _dal 02, _dal 12) Glue. X PANDA Workshop 2019 56

Usage in DSelector • You only need to do (w/ modified Make. DSelector): Make. DSelector tree_pipkmkspi 0_030300. root pipkmkspi 0_Tree pipkmkspi 0 • In DSelector_pipkmkspi 0. C uncomment / modify 3 Bool_t DSelector_pipkmkspi 0: : Init(TTree *loc. Tree). . . 107 d. Combo. Tree. Helper = new DCombo. Tree. Helper(. . . , "Ks; pi 0; pi+ K-; . . . ", . . . ); . . . 338 339 340 • // FILL FLAT TREE d. Combo. Tree. Helper->Fill(loc. Entry); Fill_Flat. Tree(); and that's it basically. . . K. Götzen Glue. X PANDA Workshop 2019 57

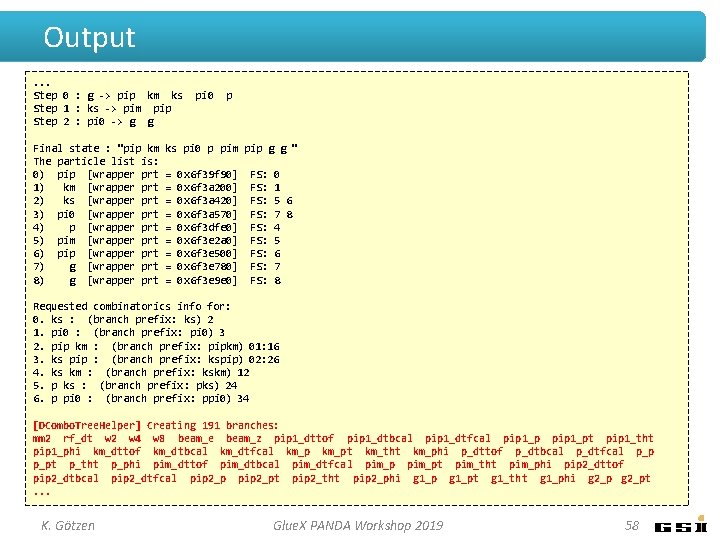
Output. . . Step 0 : g -> pip km ks Step 1 : ks -> pim pip Step 2 : pi 0 -> g g Final state : "pip km The particle list is: 0) pip [wrapper prt 1) km [wrapper prt 2) ks [wrapper prt 3) pi 0 [wrapper prt 4) p [wrapper prt 5) pim [wrapper prt 6) pip [wrapper prt 7) g [wrapper prt 8) g [wrapper prt pi 0 p ks pi 0 p pim pip g g " = = = = = 0 x 6 f 39 f 90] 0 x 6 f 3 a 200] 0 x 6 f 3 a 420] 0 x 6 f 3 a 570] 0 x 6 f 3 dfe 0] 0 x 6 f 3 e 2 a 0] 0 x 6 f 3 e 500] 0 x 6 f 3 e 780] 0 x 6 f 3 e 9 e 0] FS: FS: FS: 0 1 5 6 7 8 4 5 6 7 8 Requested combinatorics info for: 0. ks : (branch prefix: ks) 2 1. pi 0 : (branch prefix: pi 0) 3 2. pip km : (branch prefix: pipkm) 01: 16 3. ks pip : (branch prefix: kspip) 02: 26 4. ks km : (branch prefix: kskm) 12 5. p ks : (branch prefix: pks) 24 6. p pi 0 : (branch prefix: ppi 0) 34 [DCombo. Tree. Helper] Creating 191 branches: mm 2 rf_dt w 2 w 4 w 8 beam_e beam_z pip 1_dttof pip 1_dtbcal pip 1_dtfcal pip 1_pt pip 1_tht pip 1_phi km_dttof km_dtbcal km_dtfcal km_pt km_tht km_phi p_dttof p_dtbcal p_dtfcal p_pt p_tht p_phi pim_dttof pim_dtbcal pim_dtfcal pim_pt pim_tht pim_phi pip 2_dttof pip 2_dtbcal pip 2_dtfcal pip 2_pt pip 2_tht pip 2_phi g 1_pt g 1_tht g 1_phi g 2_pt. . . K. Götzen Glue. X PANDA Workshop 2019 58

Output. . . Step 0 : g -> pip km ks Step 1 : ks -> pim pip Step 2 : pi 0 -> g g Final state : "pip km The particle list is: 0) pip [wrapper prt 1) km [wrapper prt 2) ks [wrapper prt 3) pi 0 [wrapper prt 4) p [wrapper prt 5) pim [wrapper prt 6) pip [wrapper prt 7) g [wrapper prt 8) g [wrapper prt pi 0 p ks pi 0 p pim pip g g " = = = = = 0 x 6 f 39 f 90] 0 x 6 f 3 a 200] 0 x 6 f 3 a 420] 0 x 6 f 3 a 570] 0 x 6 f 3 dfe 0] 0 x 6 f 3 e 2 a 0] 0 x 6 f 3 e 500] 0 x 6 f 3 e 780] 0 x 6 f 3 e 9 e 0] FS: FS: FS: 0 1 5 6 7 8 4 5 6 7 8 Requested combinatorics info for: 0. ks : (branch prefix: ks) 2 1. pi 0 : (branch prefix: pi 0) 3 2. pip km : (branch prefix: pipkm) 01: 16 3. ks pip : (branch prefix: kspip) 02: 26 4. ks km : (branch prefix: kskm) 12 5. p ks : (branch prefix: pks) 24 6. p pi 0 : (branch prefix: ppi 0) 34 [DCombo. Tree. Helper] Creating 191 branches: mm 2 rf_dt w 2 w 4 w 8 beam_e beam_z pip 1_dttof pip 1_dtbcal pip 1_dtfcal pip 1_pt pip 1_tht pip 1_phi km_dttof km_dtbcal km_dtfcal km_pt km_tht km_phi p_dttof p_dtbcal p_dtfcal p_pt p_tht p_phi pim_dttof pim_dtbcal pim_dtfcal pim_pt pim_tht pim_phi pip 2_dttof pip 2_dtbcal pip 2_dtfcal pip 2_pt pip 2_tht pip 2_phi g 1_pt g 1_tht g 1_phi g 2_pt. . . K. Götzen Glue. X PANDA Workshop 2019 59

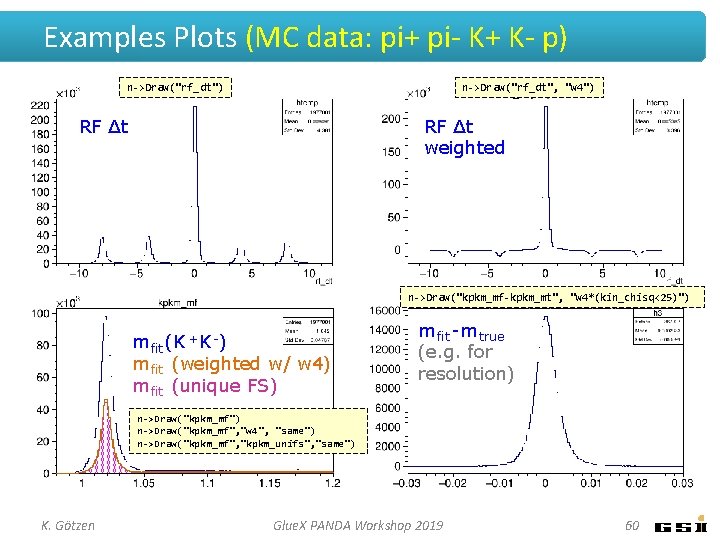
Examples Plots (MC data: pi+ pi- K+ K- p) n->Draw("rf_dt", "w 4") n->Draw("rf_dt") RF ∆t weighted RF ∆t n->Draw("kpkm_mf-kpkm_mt", "w 4*(kin_chisq<25)") m fit (K +K -) m fit (weighted w/ w 4) m fit (unique FS) m fit -m true (e. g. for resolution) n->Draw("kpkm_mf", "w 4", "same") n->Draw("kpkm_mf", "kpkm_unifs", "same") K. Götzen Glue. X PANDA Workshop 2019 60

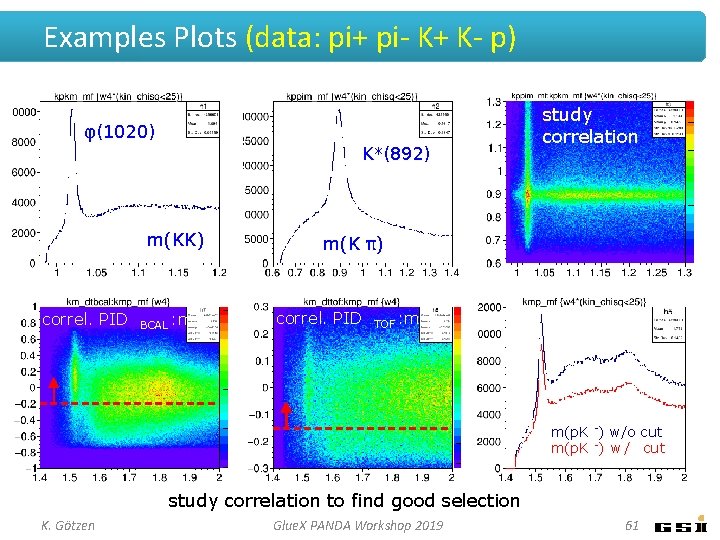
Examples Plots (data: pi+ pi- K+ K- p) φ(1020) K*(892) m(KK) correl. PID BCAL : m study correlation m(K π) correl. PID TOF : m m(p. K -) w/o cut m(p. K -) w / cut study correlation to find good selection K. Götzen Glue. X PANDA Workshop 2019 61

Summary • Panda. Root Rho – Complete framework for basic analysis – Fast simulation/stand-alone generators for quick studies – Quick Analysis / simple TTree dump allow convenient and interactive analysis work • Glue. X DSelector extension (DCombo. Tree. Helper) – Tries to transfer part of the quick analysis features – Allows to do combinatorics with no effort – Stores many quantities of general use – Could be intergrated in DSelector on github K. Götzen Glue. X PANDA Workshop 2019 62

BACKUP

PANDA Analysis Toolset - Features • Data access (Pnd. Analysis ) • Particle candidates (Rho. Candidate, Fair. Reco. Candidate • PID (Pnd. Analysis, Pnd. Pid. Combiner • Combinatorics ) ) (Rho. Cand. List ) • Particle Selectors (Rho. . . Selector, Pnd. Ana. Pid. Selector • MC truth (tree) matching ) (Pnd. Analysis: : Mc. Truth. Match ) • Fitting : POCA finder, vertex-, kinematic-, tree fitters – Pnd. Vtx. Poca, Pnd. Kalman. Vtx. Fitter, Pnd. Kin. Vtx. Fitter, Pnd. Decay. Tree. Fitter, Pnd. Kin. Fitter, Pnd 4 CFitter • N-tuples & QA Tools (Rho. Tuple, Pnd. Rho. Tuple. QA ) • Quick Analysis (Pnd. Simple. Combiner, Pnd. Simple. Combiner. Task ) • Documentation (Panda. Root Wiki) https: //panda-wiki. gsi. de/foswiki/bin/view/Computing/Panda. Root. Rho. Tutorial K. Götzen Glue. X PANDA Workshop 2019 64

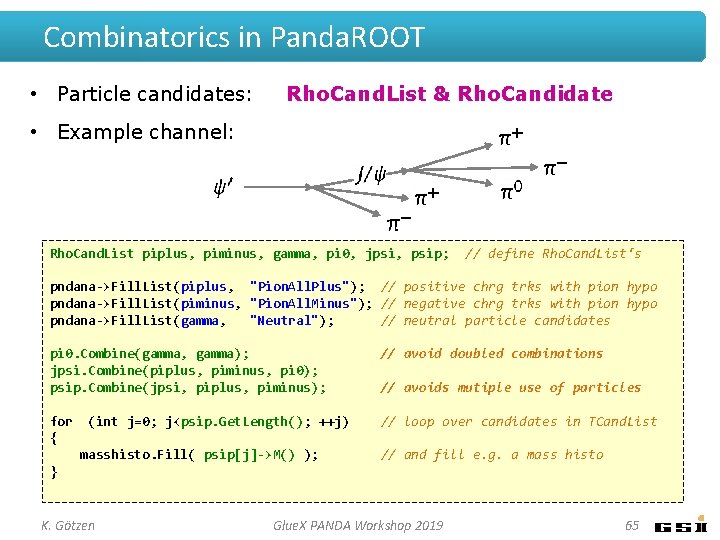
Combinatorics in Panda. ROOT • Particle candidates: Rho. Cand. List & Rho. Candidate • Example channel: Rho. Cand. List piplus, piminus, gamma, pi 0, jpsi, psip; // define Rho. Cand. List‘s pndana->Fill. List(piplus, "Pion. All. Plus"); // positive chrg trks with pion hypo pndana->Fill. List(piminus, "Pion. All. Minus"); // negative chrg trks with pion hypo pndana->Fill. List(gamma, "Neutral"); // neutral particle candidates pi 0. Combine(gamma, gamma); jpsi. Combine(piplus, piminus, pi 0); psip. Combine(jpsi, piplus, piminus); // avoid doubled combinations for { // loop over candidates in TCand. List (int j=0; j<psip. Get. Length(); ++j) masshisto. Fill( psip[j]->M() ); // avoids mutiple use of particles // and fill e. g. a mass histo } K. Götzen Glue. X PANDA Workshop 2019 65

Particle Identification

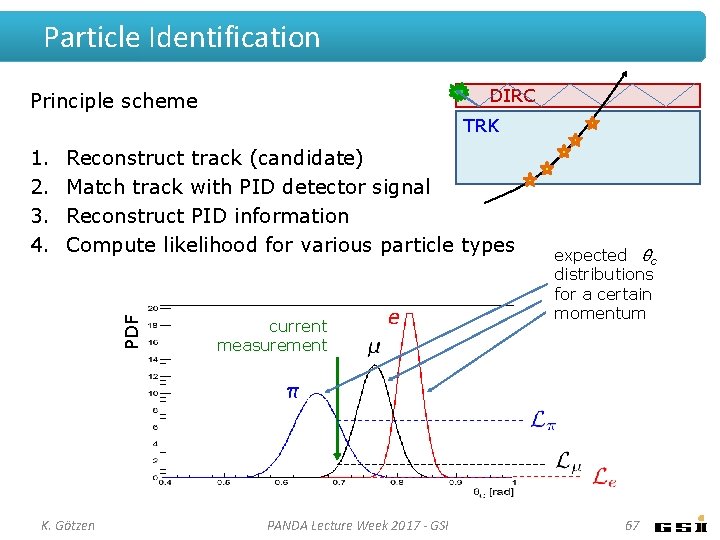
Particle Identification DIRC Principle scheme TRK Reconstruct track (candidate) Match track with PID detector signal Reconstruct PID information Compute likelihood for various particle types PDF 1. 2. 3. 4. K. Götzen current measurement PANDA Lecture Week 2017 - GSI expected θc distributions for a certain momentum 67

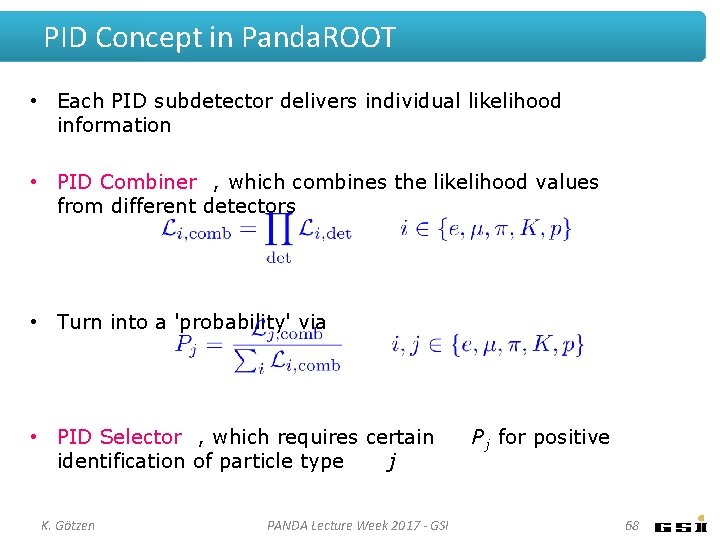
PID Concept in Panda. ROOT • Each PID subdetector delivers individual likelihood information • PID Combiner , which combines the likelihood values from different detectors • Turn into a 'probability' via • PID Selector , which requires certain identification of particle type j K. Götzen PANDA Lecture Week 2017 - GSI P j for positive 68

PID Concept in Panda. ROOT • Pnd. Analysis: : Fill. List applies PID to candidates • Predefined selection with keywords (probability based) Electron / Muon / Pion / Kaon / Proton + All / Very. Loose / Tight / Very. Tight / Best + Plus / Minus (optional) Simple keywords: Charged / Plus / Minus / Neutral / All • Available PID algorithms: Pid. Algo. Ideal. Charged Pid. Algo. Drc Pid. Algo. Ideal. Neutral Pid. Algo. Disc Pid. Algo. Mvd Pid. Algo. Stt Pid. Algo. Mdt. Hard. Cuts Pid. Algo. Emc. Bayes Pid. Algo. Sci. T Pid. Algo. Ftof Pid. Algo. Rich Pnd. Analysis *pndana = new Pnd. Analysis(); pndana->Fill. List(chrg, pndana->Fill. List(trpos, pndana->Fill. List(pipm, pndana->Fill. List(kpm, pndana->Fill. List(ep, K. Götzen "Charged"); "Plus"); "Pion. All", "Kaon. Tight", "Electron. Loose. Plus", "Pid. Algo. Mvd; Pid. Algo. Stt"); "Pid. Algo. Drd; Pid. Algo. Sci. T"); "Pid. Algo. Emc. Bayes; Pid. Algo. Mvd"); PANDA Lecture Week 2017 - GSI 69

MC Truth Matching

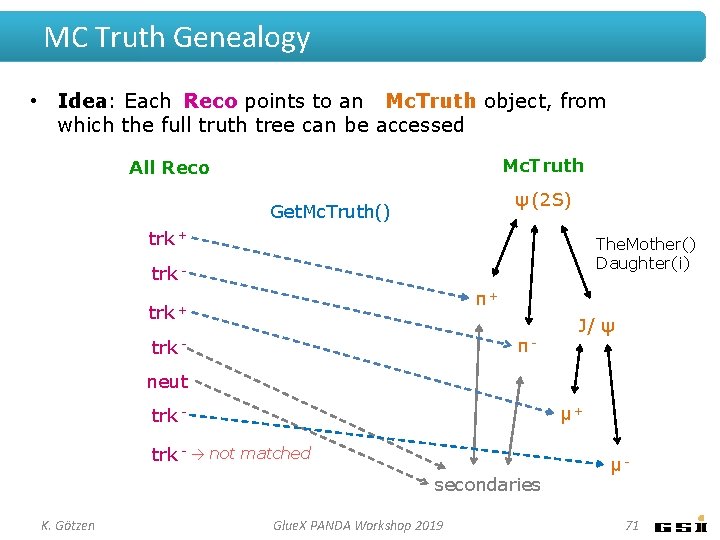
MC Truth Genealogy • Idea: Each Reco points to an Mc. Truth object, from which the full truth tree can be accessed Mc. Truth All Reco ψ (2 S) Get. Mc. Truth() trk + The. Mother() Daughter(i) trk π+ trk + π- trk - J/ ψ neut μ+ trk - → not matched secondaries K. Götzen Glue. X PANDA Workshop 2019 μ 71

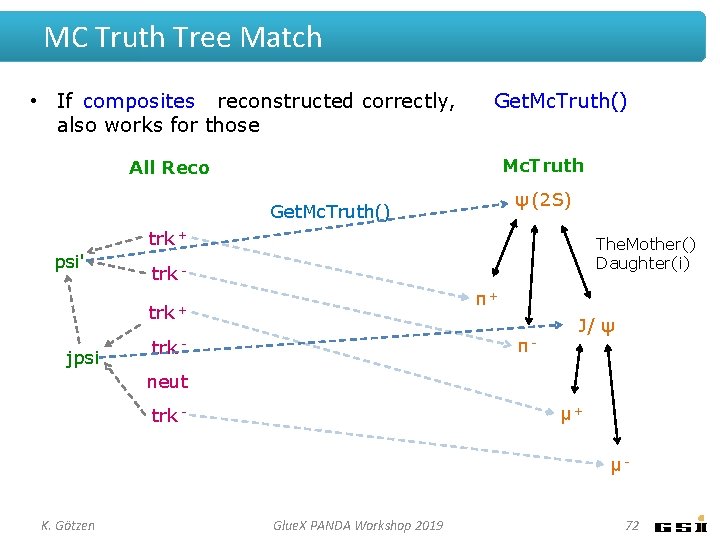
MC Truth Tree Match • If composites reconstructed correctly, also works for those Get. Mc. Truth() Mc. Truth All Reco ψ (2 S) Get. Mc. Truth() trk + psi' The. Mother() Daughter(i) trk π+ trk + jpsi π- trk - J/ ψ neut μ+ trk - μK. Götzen Glue. X PANDA Workshop 2019 72

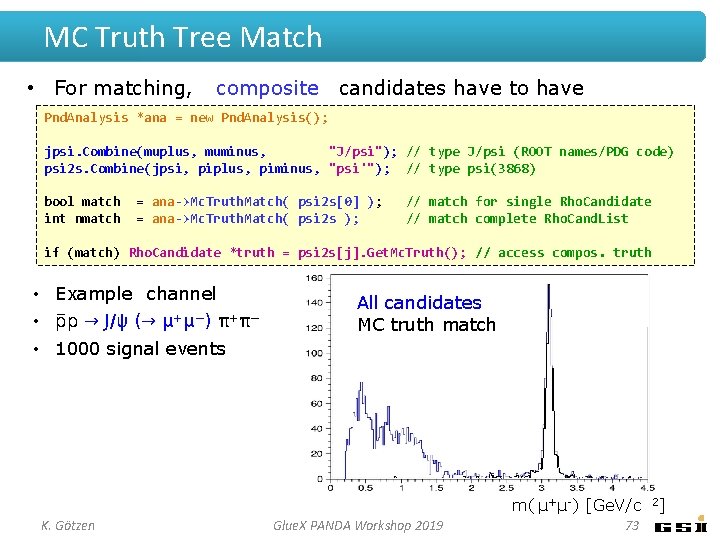
MC Truth Tree Match • For matching, composite candidates have to have type set *ana = new Pnd. Analysis(); Pnd. Analysis jpsi. Combine(muplus, muminus, "J/psi"); // type J/psi (ROOT names/PDG code) psi 2 s. Combine(jpsi, piplus, piminus, "psi'"); // type psi(3868) bool match int nmatch = ana->Mc. Truth. Match( psi 2 s[0] ); = ana->Mc. Truth. Match( psi 2 s ); // match for single Rho. Candidate // match complete Rho. Cand. List if (match) Rho. Candidate *truth = psi 2 s[j]. Get. Mc. Truth(); // access compos. truth • Example channel • p → J/ψ (→ μ+μ−) π+π− All candidates MC truth match • 1000 signal events m( μ+ μ-) [Ge. V/c K. Götzen Glue. X PANDA Workshop 2019 73 2]

Fitting

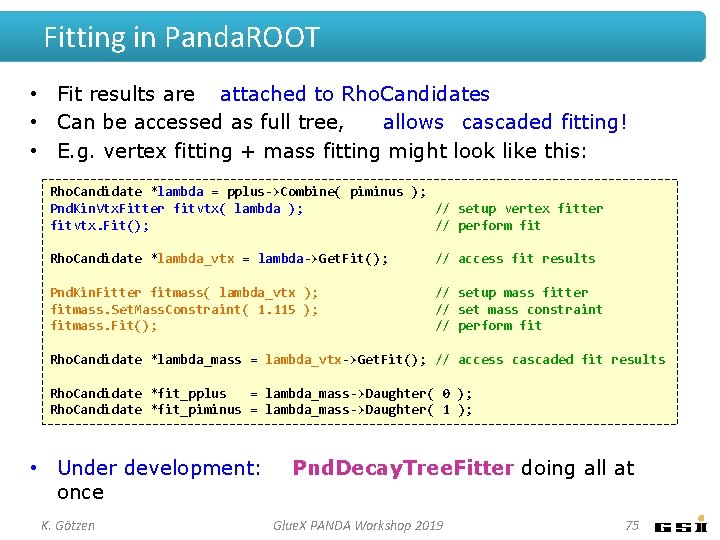
Fitting in Panda. ROOT • Fit results are attached to Rho. Candidates • Can be accessed as full tree, allows cascaded fitting! • E. g. vertex fitting + mass fitting might look like this: Rho. Candidate *lambda = pplus->Combine( piminus ); Pnd. Kin. Vtx. Fitter fitvtx( lambda ); // setup vertex fitter fitvtx. Fit(); // perform fit Rho. Candidate *lambda_vtx = lambda->Get. Fit(); // access fit results Pnd. Kin. Fitter fitmass( lambda_vtx ); fitmass. Set. Mass. Constraint( 1. 115 ); fitmass. Fit(); // setup mass fitter // set mass constraint // perform fit Rho. Candidate *lambda_mass = lambda_vtx->Get. Fit(); // access cascaded fit results Rho. Candidate *fit_pplus = lambda_mass->Daughter( 0 ); Rho. Candidate *fit_piminus = lambda_mass->Daughter( 1 ); • Under development: once K. Götzen Pnd. Decay. Tree. Fitter doing all at Glue. X PANDA Workshop 2019 75

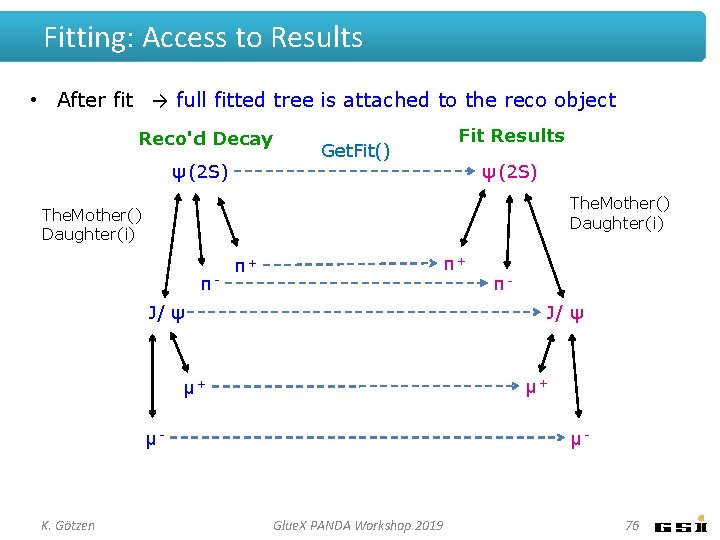
Fitting: Access to Results • After fit → full fitted tree is attached to the reco object Reco'd Decay ψ (2 S) Get. Fit() Fit Results ψ (2 S) The. Mother() Daughter(i) π- π+ π+ J/ ψ μ+ μ+ μ- K. Götzen π- μ- Glue. X PANDA Workshop 2019 76

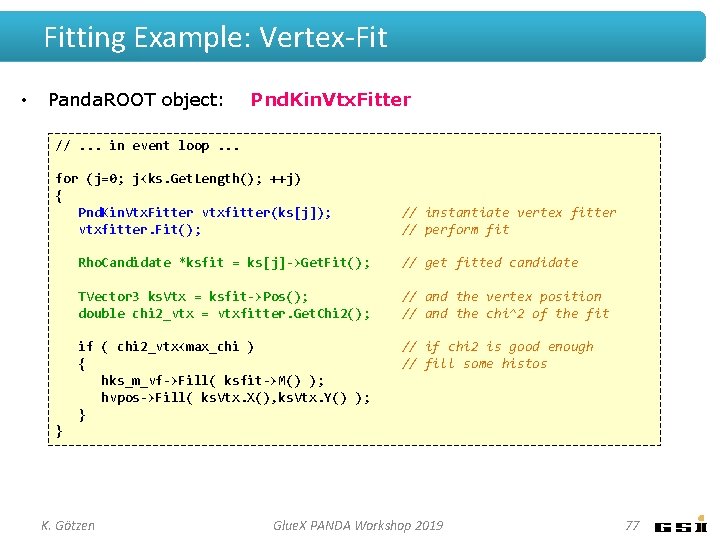
Fitting Example: Vertex-Fit • Panda. ROOT object: Pnd. Kin. Vtx. Fitter //. . . in event loop. . . for (j=0; j<ks. Get. Length(); ++j) { Pnd. Kin. Vtx. Fitter vtxfitter(ks[j]); vtxfitter. Fit(); // instantiate vertex fitter // perform fit Rho. Candidate *ksfit = ks[j]->Get. Fit(); // get fitted candidate TVector 3 ks. Vtx = ksfit->Pos(); double chi 2_vtx = vtxfitter. Get. Chi 2(); // and the vertex position // and the chi^2 of the fit if ( chi 2_vtx<max_chi ) { hks_m_vf->Fill( ksfit->M() ); hvpos->Fill( ks. Vtx. X(), ks. Vtx. Y() ); } // if chi 2 is good enough // fill some histos } K. Götzen Glue. X PANDA Workshop 2019 77

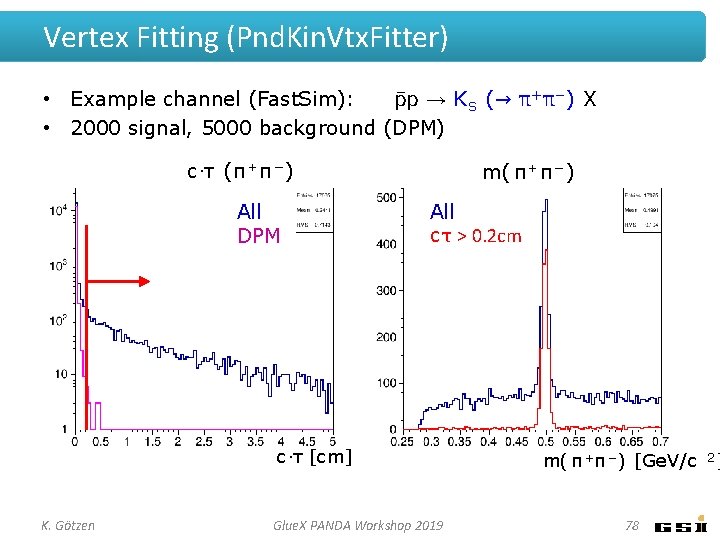
Vertex Fitting (Pnd. Kin. Vtx. Fitter) • Example channel (Fast. Sim): p → K S (→ π+π−) X • 2000 signal, 5000 background (DPM) c·τ (π + π − ) All DPM m( π + π − ) All c τ > 0. 2 cm c·τ [cm] K. Götzen Glue. X PANDA Workshop 2019 m( π +π −) [Ge. V/c 78 2]

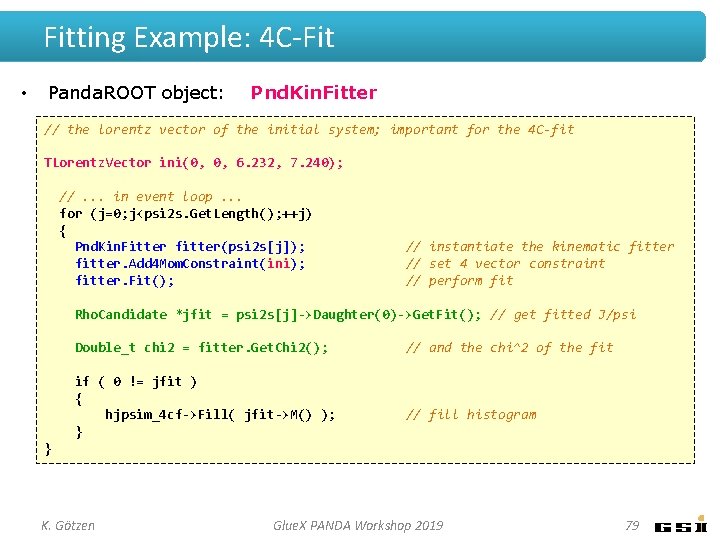
Fitting Example: 4 C-Fit • Panda. ROOT object: Pnd. Kin. Fitter // the lorentz vector of the initial system; important for the 4 C-fit TLorentz. Vector ini(0, 0, 6. 232, 7. 240); //. . . in event loop. . . for (j=0; j<psi 2 s. Get. Length(); ++j) { Pnd. Kin. Fitter fitter(psi 2 s[j]); fitter. Add 4 Mom. Constraint(ini); fitter. Fit(); // instantiate the kinematic fitter // set 4 vector constraint // perform fit Rho. Candidate *jfit = psi 2 s[j]->Daughter(0)->Get. Fit(); // get fitted J/psi Double_t chi 2 = fitter. Get. Chi 2(); if ( 0 != jfit ) { hjpsim_4 cf->Fill( jfit->M() ); } // and the chi^2 of the fit // fill histogram } K. Götzen Glue. X PANDA Workshop 2019 79

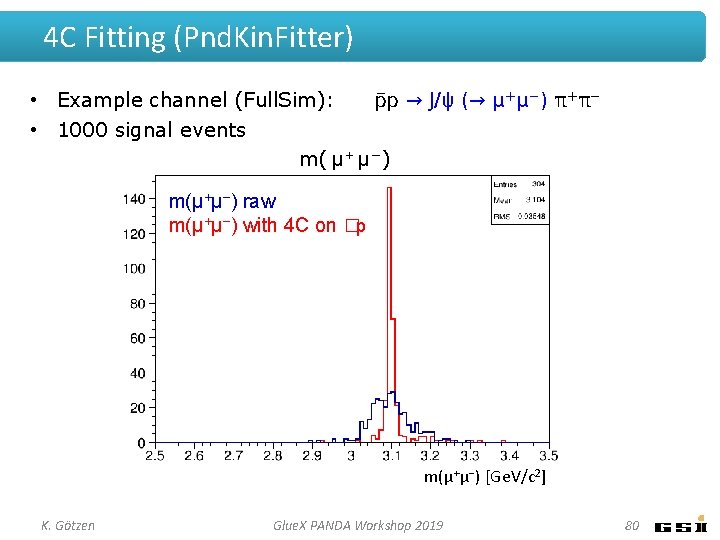
4 C Fitting (Pnd. Kin. Fitter) • Example channel (Full. Sim): p → J/ψ (→ μ+μ−) π+π− • 1000 signal events m( μ + μ − ) m(μ+μ−) raw m(μ+μ−) with 4 C on �p m(μ+μ−) [Ge. V/c 2] K. Götzen Glue. X PANDA Workshop 2019 80

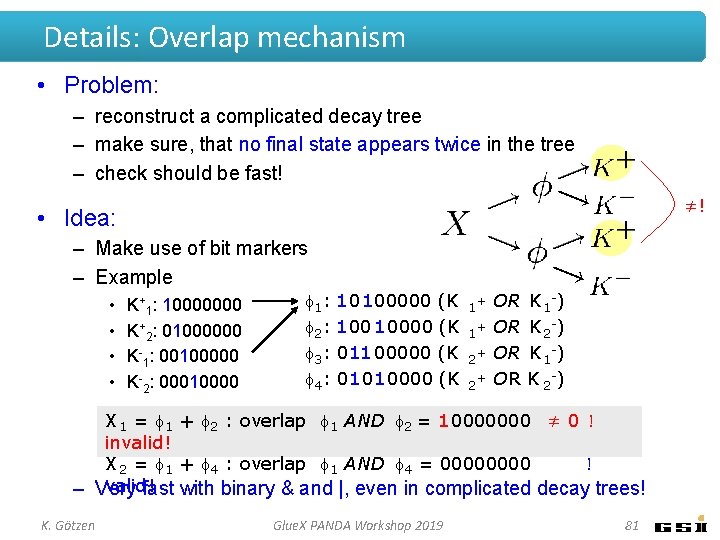
Details: Overlap mechanism • Problem: – reconstruct a complicated decay tree – make sure, that no final state appears twice in the tree – check should be fast! ≠! • Idea: – Make use of bit markers – Example • • K+1: 10000000 K+2: 01000000 K-1: 00100000 K-2: 00010000 1 : 10100000 (K 2 : 10000 (K 3 : 011 00000 (K 4 : 01010000 (K 1 1 2 2 + + OR OR K 1 -) K 2 -) X 1 = 1 + 2 : overlap 1 AND 2 = 10000000 ≠ 0 ! invalid! X 2 = 1 + 4 : overlap 1 AND 4 = 0000 ! valid! – Very fast with binary & and |, even in complicated decay trees! K. Götzen Glue. X PANDA Workshop 2019 81

PID Concept in Panda. ROOT: Pnd. Ana. Pid. Combiner, Pnd. Ana. Pid. Selector • Pnd. Ana. Pid. Combiner – combines on demand probabilities from various algorithms by computing product of all P k (k=algorithms) – copies resulting probabilities to Rho. Candidate/Rho. Cand. List • Pnd. Ana. Pid. Selector – selects particles based on these probabilities • Pnd. Analysis: : Fill. List is a short-cut to this funtionallity via pndana. Fill. List(list, “Electron. Loose“, "Pid. Algo. Emc. Bayes; Pid. Algo. Drc"); • Predefined selection with keywords (probability based) Electron / Muon / Pion / Kaon / Proton + All / Very. Loose / Tight / Very. Tight / Best + Plus / Minus (optional) Simple keywords Charged / Plus / Minus / Neutral / All K. Götzen PANDA Lecture Week 2017 - GSI 82

PID Concept in Panda. ROOT • Panda. ROOT: Pnd. Analysis, Pnd. Ana. Pid. Combiner, Pnd. Ana. Pid. Selector Pnd. Analysis *pndana= new Pnd. Analysis(); pndana->Fill. List(eplus, "Electron. Loose. Plus", "Pid. Algo. Emc. Bayes; Pid. Algo. Mvd"); pndana->Fill. List(eminus, "Electron. Loose. Minus", "Pid. Algo. Emc. Bayes; Pid. Algo. Mvd"); Or 'by hand': Rho. Cand. List charged, kaon. Loose; Pnd. Ana. Pid. Selector kaon. Sel("Kaon. Selector"); kaon. Sel. Set. Selection("Kaon. Loose"); // set selection criterion Pnd. Ana. Pid. Combiner pid. Comb("Pid. Combiner"); pid. Comb. Set. Tca. Names("Pid. Algo. Drc; Pid. Algo. Mvd"); // set algo‘s while (evr->Get. Event()) { pndana->Fill. List(charged, "Charged"); // start w/ charged candidates pid. Comb. Apply(charged); kaon. Loose. Select(charged, kaon. Sel); // copy probab. to candidates // select kaons from charged } K. Götzen PANDA Lecture Week 2017 - GSI 83

Pnd. Rho. Tuple. QA - TTree made simple • Provides QA functions for persisting values in TTree // *** QA for candidates void qa. Cand(TString pre, Rho. Candidate *cc, Rho. Tuple *n, bool skip=false); void qa. P 4(TString pre, TLorentz. Vector c, Rho. Tuple *n, bool skip=false); void qa. P 4 Cms(TString pre, TLorentz. Vector c, Rho. Tuple *n, bool skip=false); void qa. P 4 Cov(TString pre, Rho. Candidate *c, Rho. Tuple *n, bool skip=false); // *** QA for composites void qa. Comp(TString pre, Rho. Candidate *c, Rho. Tuple *n); void qa. Ks 0(TString pre, Rho. Candidate *c, Rho. Tuple *n); void qa. Pi 0(TString pre, Rho. Candidate *c, Rho. Tuple *n); Very handy function to store composite info! // *** QA of event shape void qa. Event. Shape(TString pre, Pnd. Event. Shape *evsh, Rho. Tuple *n); void qa. Event. Shape. Short(TString pre, Pnd. Event. Shape *evsh, Rho. Tuple *n); // *** QA for parts of eventshape void qa. ESPid. Mult(TString pre, Pnd. Event. Shape *evsh, double prob, double pmin, Rho. Tuple *n); void qa. ESMult(TString pre, Pnd. Event. Shape *evsh, Rho. Tuple *n); void qa. ESSum(TString pre, Pnd. Event. Shape *evsh, Rho. Tuple *n); void qa. ESMin. Max(TString pre, Pnd. Event. Shape *evsh, Rho. Tuple *n); void qa. ESEvent. Vars(TString pre, Pnd. Event. Shape *evsh, Rho. Tuple *n); // *** QA track, vtx, PID, decay void qa. Vtx(TString pre, Rho. Candidate *c, Rho. Tuple *n); void qa. Poca(TString pre, Rho. Candidate *c, Rho. Tuple *n ); . . K. Götzen Glue. X PANDA Workshop 2019 84

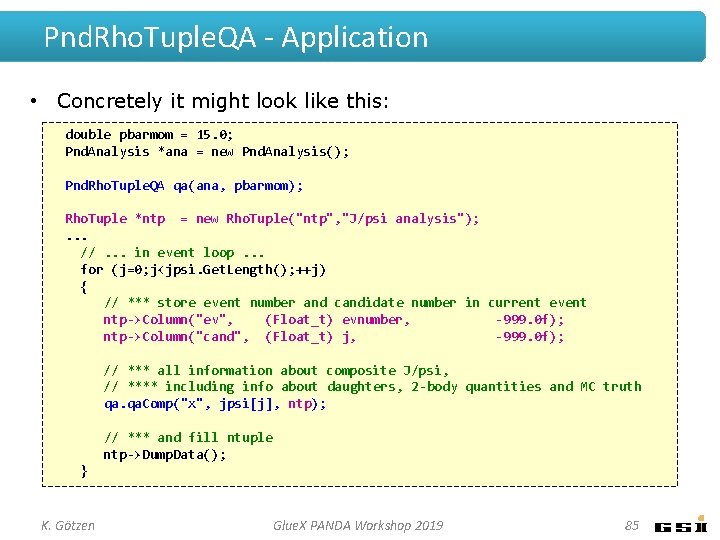
Pnd. Rho. Tuple. QA - Application • Concretely it might look like this: double pbarmom = 15. 0; Pnd. Analysis *ana = new Pnd. Analysis(); Pnd. Rho. Tuple. QA qa(ana, pbarmom); Rho. Tuple *ntp = new Rho. Tuple("ntp", "J/psi analysis"); . . . //. . . in event loop. . . for (j=0; j<jpsi. Get. Length(); ++j) { // *** store event number and candidate number in current event ntp->Column("ev", (Float_t) evnumber, -999. 0 f); ntp->Column("cand", (Float_t) j, -999. 0 f); // *** all information about composite J/psi, // **** including info about daughters, 2 -body quantities and MC truth qa. Comp("x", jpsi[j], ntp); // *** and fill ntuple ntp->Dump. Data(); } K. Götzen Glue. X PANDA Workshop 2019 85

Fast Simulation Details

Detector Modules Target Spectrometer TRK EMC PID K. Götzen STT Forward Spectrometer STT + MVD + GEM EMC Barrel EMC Fwd TS MVD Vtx EMC FS EMC Bwd EMC Barrel STT PID DIRC PID EMC FS PID EMC Fwd MVD PID DISC PID RICH PID EMC Bwd MUO PID Glue. X PANDA Workshop 2019 MUO FS PID 87

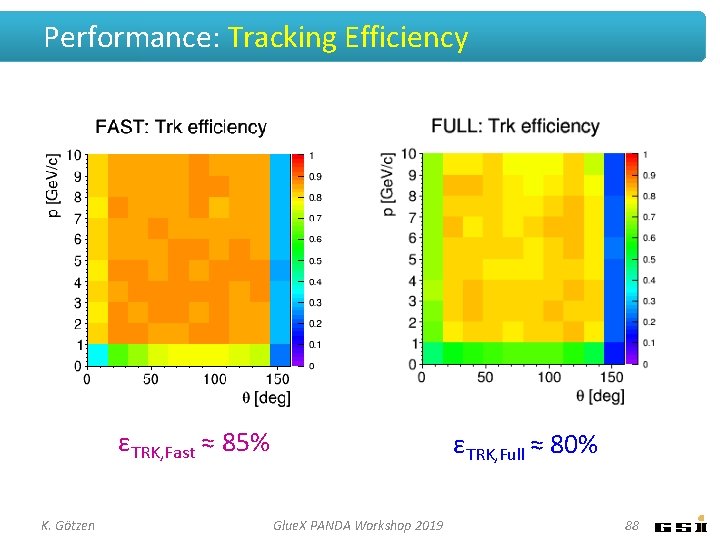
Performance: Tracking Efficiency εTRK, Fast ≈ 85% K. Götzen εTRK, Full ≈ 80% Glue. X PANDA Workshop 2019 88

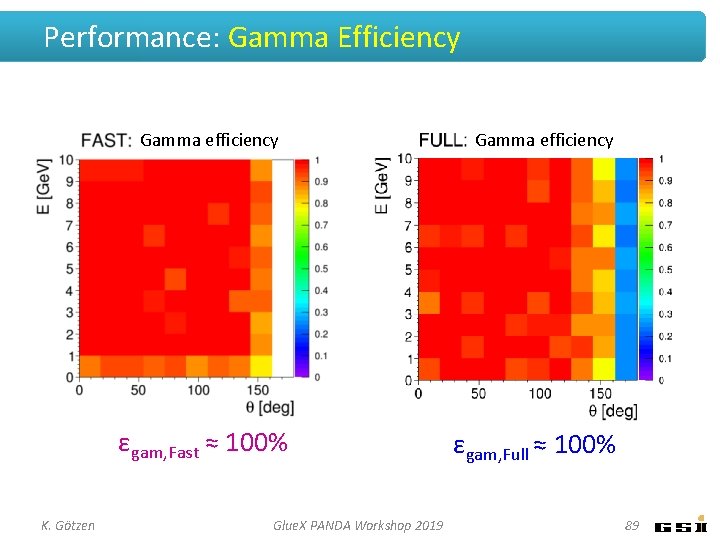
Performance: Gamma Efficiency Gamma efficiency εgam, Fast ≈ 100% K. Götzen Glue. X PANDA Workshop 2019 Gamma efficiency εgam, Full ≈ 100% 89

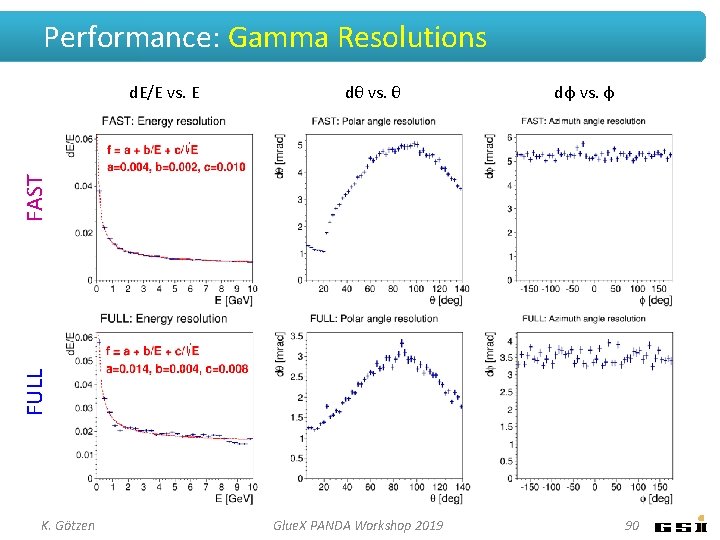
Performance: Gamma Resolutions dθ vs. θ dφ vs. φ FULL FAST d. E/E vs. E K. Götzen Glue. X PANDA Workshop 2019 90

Event Filtering in Panda. Root Pnd. Filtered. Primary. Generator (w/ Pnd. Smp. Filt, Pnd. Smp. Cand) • Complete Normal Form (DNF = logic via , Disjunctive e. g. ) • Filter on: Multiplicity, kinematic quantities, composite mass • Filterset: Individual filters combined with ' &&' (AND) • Multiple filtersets combined with '||' (OR) • Negate individual filters with '!' (NOT) • Example: Pnd. Filtered. Primary. Generator* prim. Gen Pnd. Filtered. Primary. Generator(); !N trk ≤ 3 AND N gam ≤ 6 AND= new N(2. 0<M(e+ e-)<4. 0 Ge. V/c 2 ) ≥ 1 prim. Gen->Add. Filter("!(t+; . . 3) && (gam ; . . 6) && M(e+ e- ; m[3. 0, 2. 0])"); Aug. 14, 2018 K. Götzen - Panda. Root Meeting (No number range given defaults to '1. . ' = at least one) 91
![Filter Syntax Counts particle mult kinematic cuts particle particle type MC Filter Syntax: • Counts: "(<particle> ; [<mult>] ; [kinematic cuts])" <particle>: particle type (MC](https://slidetodoc.com/presentation_image_h2/5a413baab1f8a3cddf628100ce512ee5/image-92.jpg)
Filter Syntax: • Counts: "(<particle> ; [<mult>] ; [kinematic cuts])" <particle>: particle type (MC truth) to count any (tracks/neutrals), t+- (tracks), t+, t-, nt/gam (neutrals) e+, e-, e+-, mu+, mu-, mu+-, pi+, pi-, pi+-, k+, k-, k+p+, p-, p+-, n 0 (neutron), n 0 b, n 00 b (neutron or anti-neutron) pdg[<pdg code>], pdg[<pdg code>*1000] (incl. anti-particle) • Inv. mass: "M(<p 1> <p 2> [<p 3>. . <p 5>] ; m[ctr, wid] ; [<mult>] ; [kin. cuts])" <pi> : particle hypothesis to be applied for combinatorics e+, e-, mu+, mu-, pi+, pi-, k+, k-, p+, p-, gam m[ctr, wid] : mass window | minv - ctr | ≤ wid / 2 Aug. 14, 2018 K. Götzen - Panda. Root Meeting 92

Filter Syntax: • <mult>: multiplicity short cuts examples min. . max : min ≤ N ≤ max num = num. . max = 0. . max min. . = min. . 10000 none = 1. . 10000 (t+- ; 2. . 4) (pdg[3122] ; 1) (nt ; . . 2) (any ; 4. . ) (K+) • <kinematic cuts>: optional cuts on p, pt, pz, tht, phi (= keyword X) p, pt, and pz in [Ge. V/c] tht and phi in [deg] short cuts Aug. 14, 2018 X[min, max] : min ≤ X ≤ max X[ , max] = X[0 , max] X[min, ] = X[min, 1 E 8] K. Götzen - Panda. Root Meeting examples (t+- ; phi[5, 10]) (nt ; tht[, 20]) (mu+- ; pt[1, ]) 93

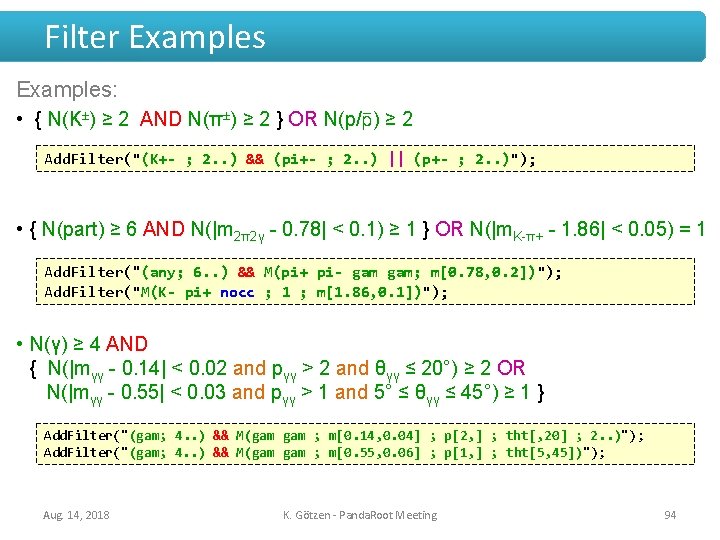
Filter Examples: • { N(K±) ≥ 2 AND N(π±) ≥ 2 } OR N(p/ ) ≥ 2 Add. Filter("(K+- ; 2. . ) && (pi+- ; 2. . ) || (p+- ; 2. . )"); • { N(part) ≥ 6 AND N(|m 2π2γ - 0. 78| < 0. 1) ≥ 1 } OR N(|m. K-π+ - 1. 86| < 0. 05) = 1 Add. Filter("(any; 6. . ) && M(pi+ pi- gam; m[0. 78, 0. 2])"); Add. Filter("M(K- pi+ nocc ; 1 ; m[1. 86, 0. 1])"); • N(γ) ≥ 4 AND { N(|mγγ - 0. 14| < 0. 02 and pγγ > 2 and θγγ ≤ 20°) ≥ 2 OR N(|mγγ - 0. 55| < 0. 03 and pγγ > 1 and 5° ≤ θγγ ≤ 45°) ≥ 1 } Add. Filter("(gam; 4. . ) && M(gam ; m[0. 14, 0. 04] ; p[2, ] ; tht[, 20] ; 2. . )"); Add. Filter("(gam; 4. . ) && M(gam ; m[0. 55, 0. 06] ; p[1, ] ; tht[5, 45])"); Aug. 14, 2018 K. Götzen - Panda. Root Meeting 94