Analysis of Sorting by Transpositions based on Algebraic
Analysis of Sorting by Transpositions based on Algebraic Formalism Cleber V. G. Mira João Meidanis RECOMB 2004
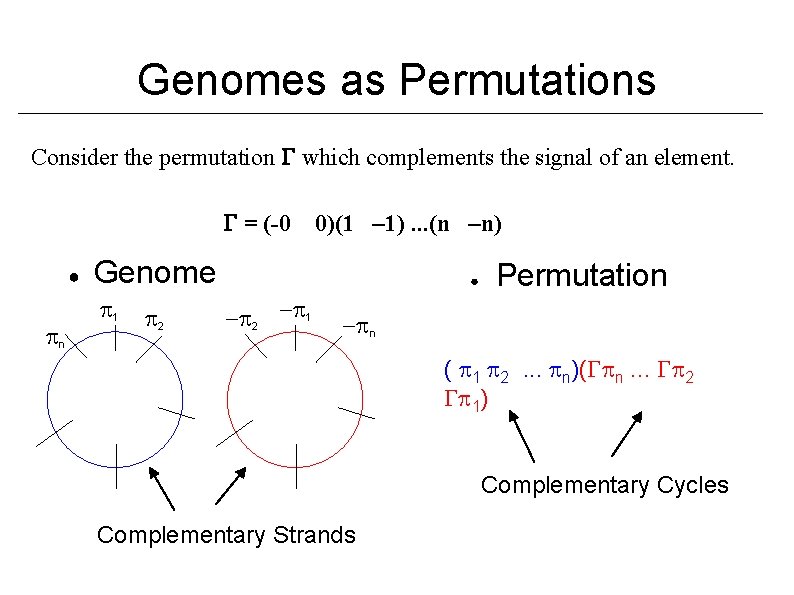
Genomes as Permutations Consider the permutation which complements the signal of an element. = (-0 ● n 0)(1 – 1). . . (n –n) Genome 1 2 ● - 2 - 1 Permutation - n ( 1 2. . . n)( n. . . 2 1) Complementary Cycles Complementary Strands
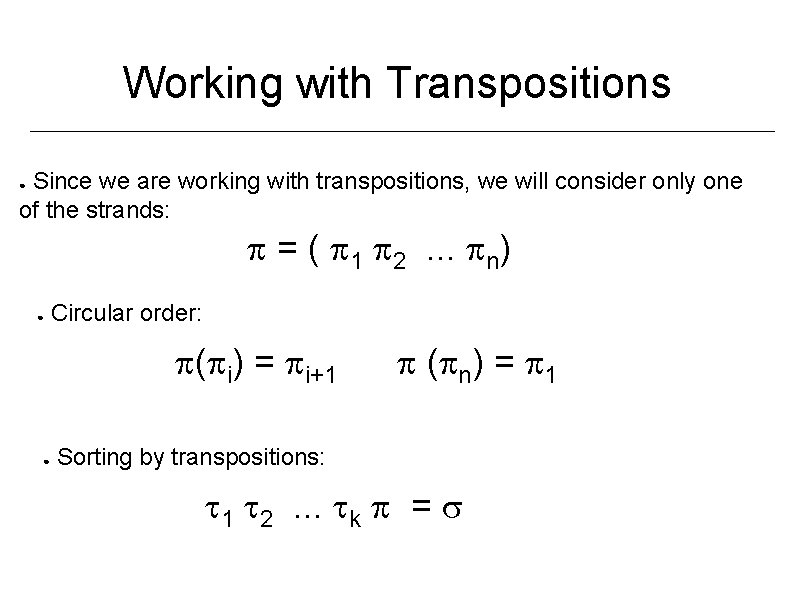
Working with Transpositions Since we are working with transpositions, we will consider only one of the strands: ● = ( 1 2. . . n) ● Circular order: ( i) = i+1 ● ( n) = 1 Sorting by transpositions: 1 2. . . k =
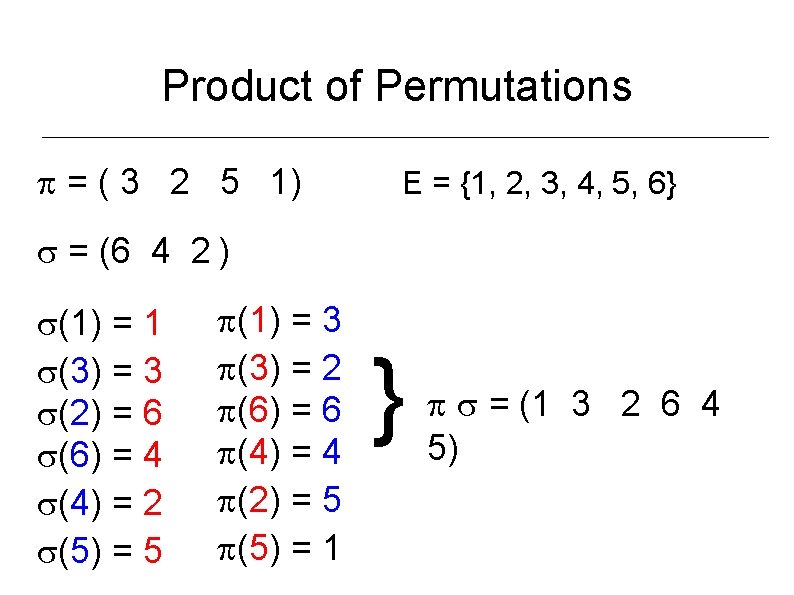
Product of Permutations = ( 3 2 5 1) E = {1, 2, 3, 4, 5, 6} = (6 4 2 ) (1) = 1 (3) = 3 (2) = 6 (6) = 4 (4) = 2 (5) = 5 (1) = 3 (3) = 2 (6) = 6 (4) = 4 (2) = 5 (5) = 1 } = (1 3 2 6 4 5)
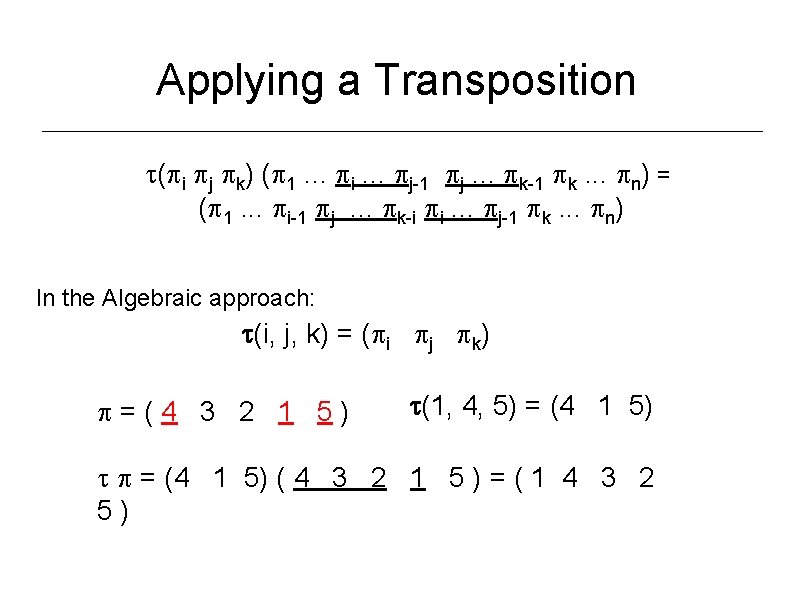
Applying a Transposition ( i j k) ( 1. . . i. . . j-1 j. . . k-1 k. . . n) = ( 1. . . i-1 j. . . k-i i. . . j-1 k. . . n) In the Algebraic approach: (i, j, k) = ( i j k) =(4 3 2 1 5) (1, 4, 5) = (4 1 5) ( 4 3 2 1 5 ) = ( 1 4 3 2 5)

2 -cycle Decomposition ● Every permutation has a 2 -cycle decomposition. = (4 3 2 1 5) = (4 3)(3 2)(2 1)(1 5) ● ● Odd cycles have an even number of 2 cycles in their 2 -cycle decomposition. The norm of is the minimum number of cycles in the 2 -cycle decomposition of .
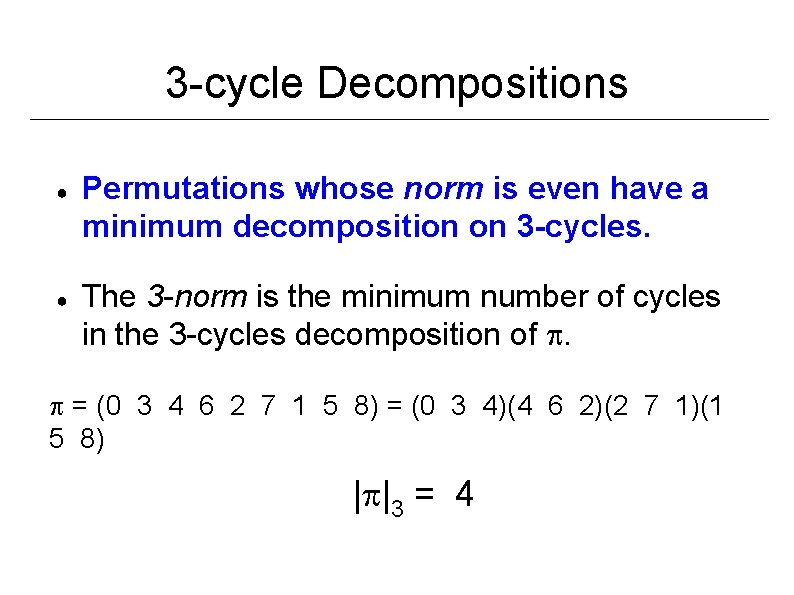
3 -cycle Decompositions ● ● Permutations whose norm is even have a minimum decomposition on 3 -cycles. The 3 -norm is the minimum number of cycles in the 3 -cycles decomposition of . = (0 3 4 6 2 7 1 5 8) = (0 3 4)(4 6 2)(2 7 1)(1 5 8) | |3 = 4
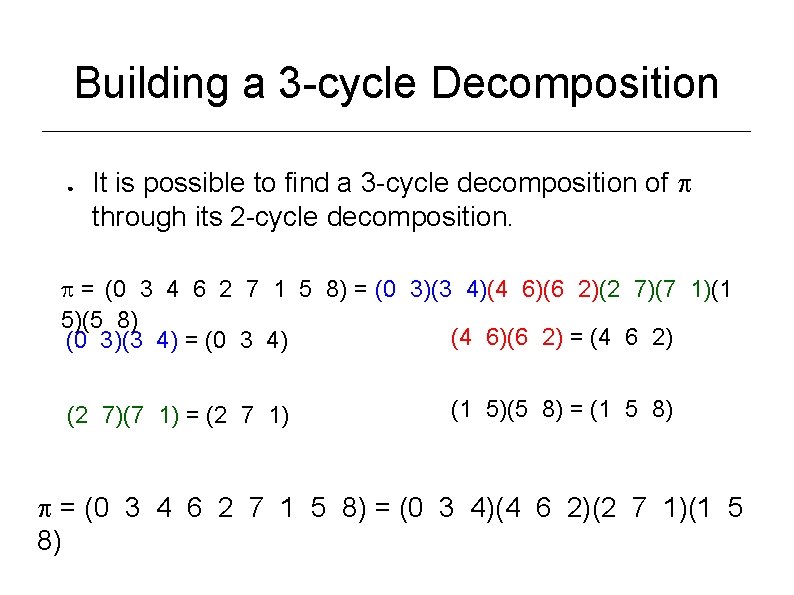
Building a 3 -cycle Decomposition ● It is possible to find a 3 -cycle decomposition of through its 2 -cycle decomposition. = (0 3 4 6 2 7 1 5 8) = (0 3)(3 4)(4 6)(6 2)(2 7)(7 1)(1 5)(5 8) (4 6)(6 2) = (4 6 2) (0 3)(3 4) = (0 3 4) (2 7)(7 1) = (2 7 1) (1 5)(5 8) = (1 5 8) = (0 3 4 6 2 7 1 5 8) = (0 3 4)(4 6 2)(2 7 1)(1 5 8)
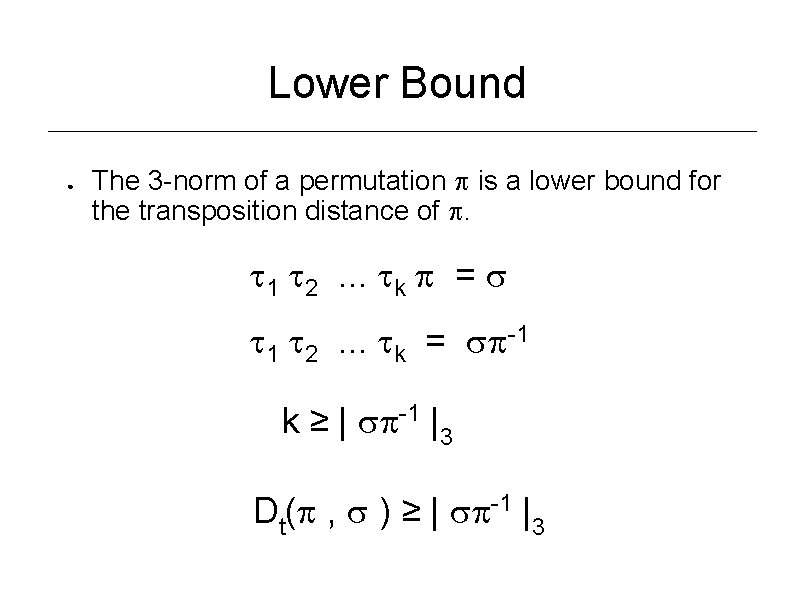
Lower Bound ● The 3 -norm of a permutation is a lower bound for the transposition distance of . 1 2. . . k = 1 2. . . k = -1 k ≥ | -1 |3 Dt( , ) ≥ | -1 |3
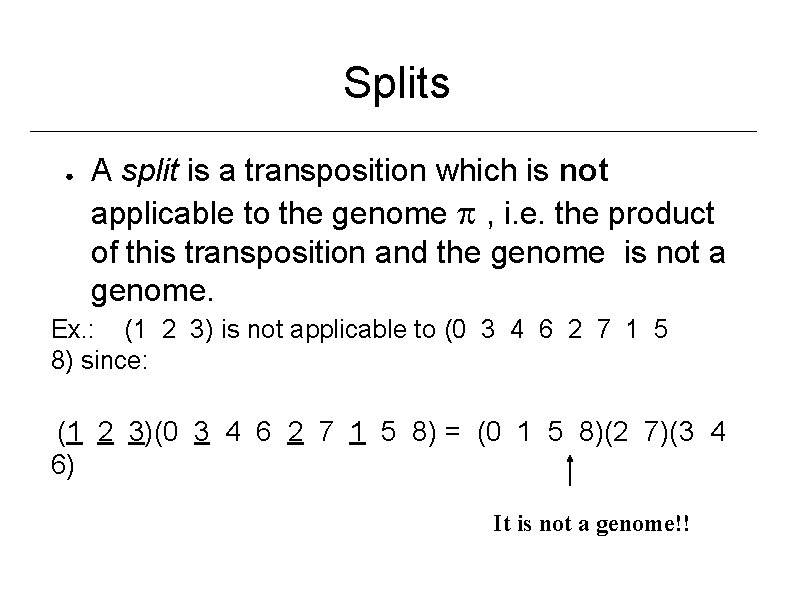
Splits ● A split is a transposition which is not applicable to the genome , i. e. the product of this transposition and the genome is not a genome. Ex. : (1 2 3) is not applicable to (0 3 4 6 2 7 1 5 8) since: (1 2 3)(0 3 4 6 2 7 1 5 8) = (0 1 5 8)(2 7)(3 4 6) It is not a genome!!
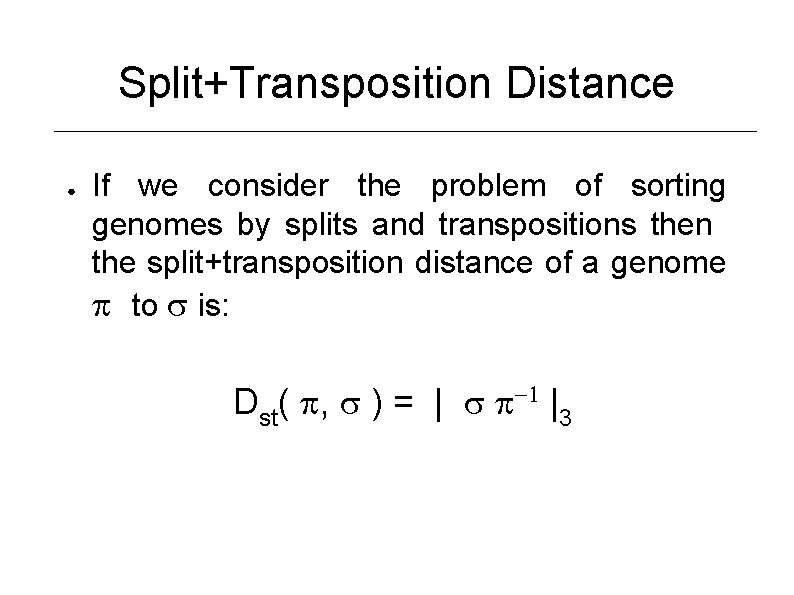
Split+Transposition Distance ● If we consider the problem of sorting genomes by splits and transpositions then the split+transposition distance of a genome to is: Dst( , ) = | -1 |3

Bibliography ● ● V. Bafna and P. A. Pevzner, 1995, Sorting by Transpositions. In: “Proceedings of the Sixth Annual ACM-SIAM Symposium on Discrete Algorithms”, San Francisco, USA, pp. 614 -623 J. Meidanis and Z. Dias, 2000, An Alternative Algebraic Formalism for Genome Rearrangements. In: “Comparative Genomics: Empirical and Analytical Approaches to Gene Order Dynamics, Map Alignment and Evolution of Gene Families” D. Sankoff and J. H. Nadeau, editors, pp. 213 -223
- Slides: 12