Analysis of Multiple Experiments TIGR Multiple Experiment Viewer



- Slides: 16

Analysis of Multiple Experiments TIGR Multiple Experiment Viewer (Me. V) Joseph White DFCI January 24, 2008

Me. V • • Stand-alone java application for analysis New version: 4. 1 Not database centric; uses TDMS files Writes TDMS files Primarily for normalized data Me. V does not currently write MAGE-TAB Download Me. V from: tm 4. org

Outline • • Description of Me. V How Me. V treats expression Some essential concepts Demo: basic operations in Me. V – New file loader – ANOVA example • Demo of Me. V new features – Affymetrix file reader – Non-parametric tests – CGH • GCOD

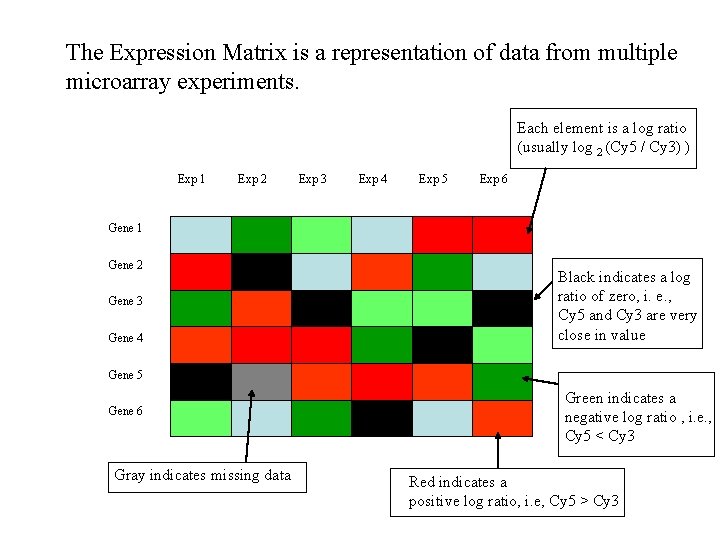
The Expression Matrix is a representation of data from multiple microarray experiments. Each element is a log ratio (usually log 2 (Cy 5 / Cy 3) ) Exp 1 Exp 2 Exp 3 Exp 4 Exp 5 Exp 6 Gene 1 Gene 2 Gene 3 Gene 4 Black indicates a log ratio of zero, i. e. , Cy 5 and Cy 3 are very close in value Gene 5 Gene 6 Gray indicates missing data Green indicates a negative log ratio , i. e. , Cy 5 < Cy 3 Red indicates a positive log ratio, i. e, Cy 5 > Cy 3

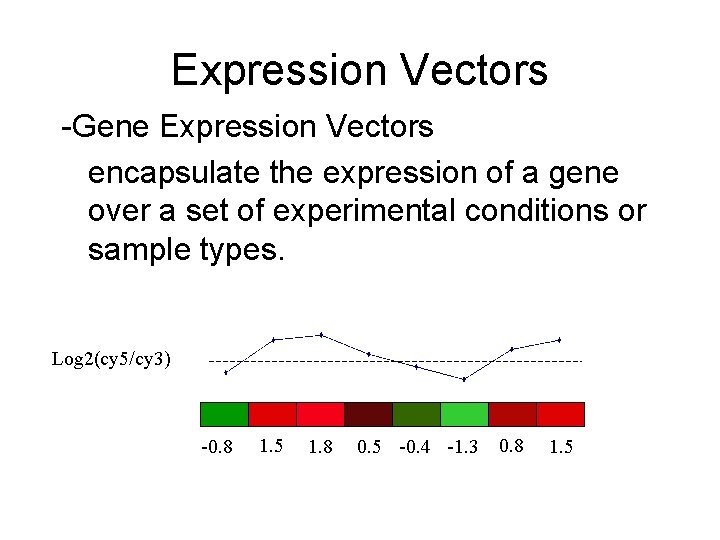
Expression Vectors -Gene Expression Vectors encapsulate the expression of a gene over a set of experimental conditions or sample types. Log 2(cy 5/cy 3) -0. 8 1. 5 1. 8 0. 5 -0. 4 -1. 3 0. 8 1. 5

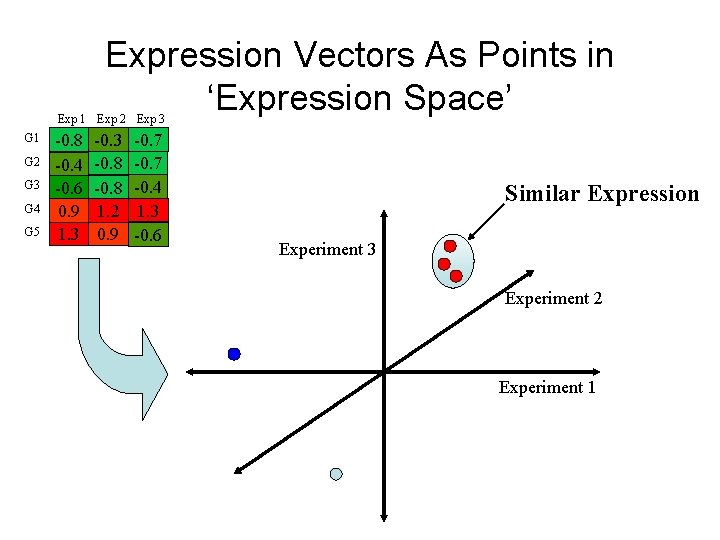
Expression Vectors As Points in ‘Expression Space’ Exp 1 Exp 2 Exp 3 G 1 G 2 G 3 G 4 G 5 -0. 8 -0. 4 -0. 6 0. 9 1. 3 -0. 8 1. 2 0. 9 -0. 7 -0. 4 1. 3 -0. 6 Similar Expression Experiment 3 Experiment 2 Experiment 1

Distance and Similarity -the ability to calculate a distance (or similarity, it’s inverse) between two expression vectors is fundamental to clustering algorithms -distance between vectors is the basis upon which decisions are made when grouping similar patterns of expression -selection of a distance metric defines the concept of distance

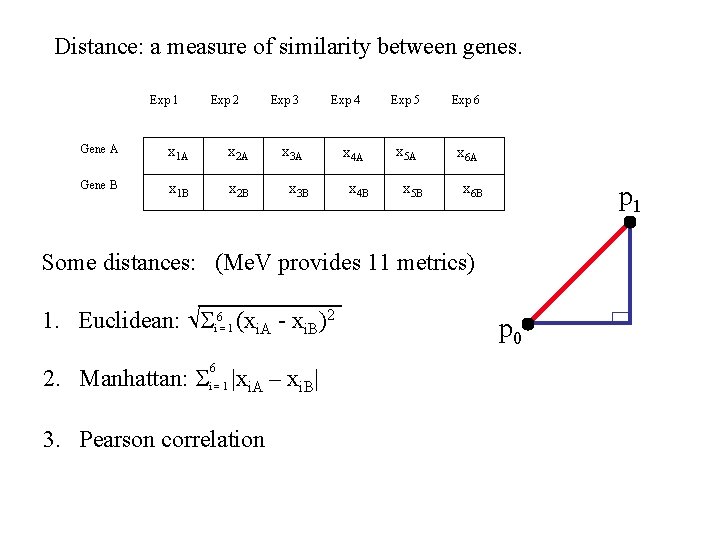
Distance: a measure of similarity between genes. Exp 1 Exp 2 Gene A x 1 A x 2 A Gene B x 1 B x 2 B Exp 3 Exp 4 x 3 A x 3 B x 4 A x 4 B Exp 5 Exp 6 x 5 A x 6 A x 5 B x 6 B p 1 Some distances: (Me. V provides 11 metrics) 1. Euclidean: i 6= 1 (xi. A - xi. B)2 6 2. Manhattan: i = 1 |xi. A – xi. B| 3. Pearson correlation p 0

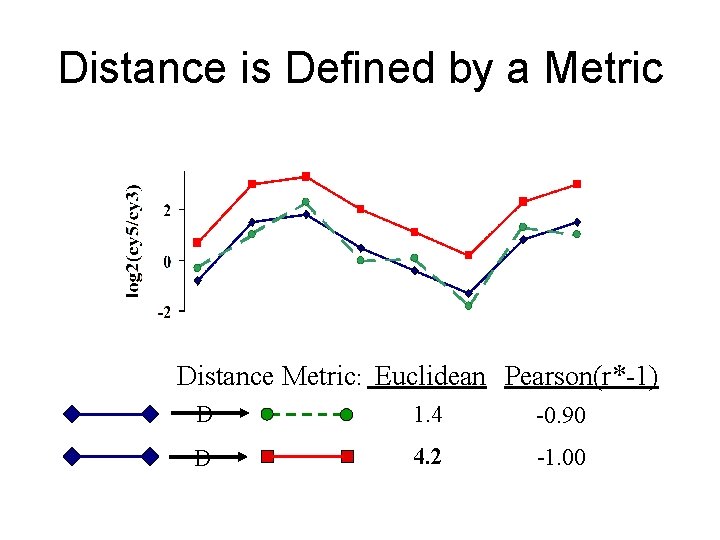
Distance is Defined by a Metric Distance Metric: Euclidean Pearson(r*-1) D 1. 4 -0. 90 D 4. 2 -1. 00

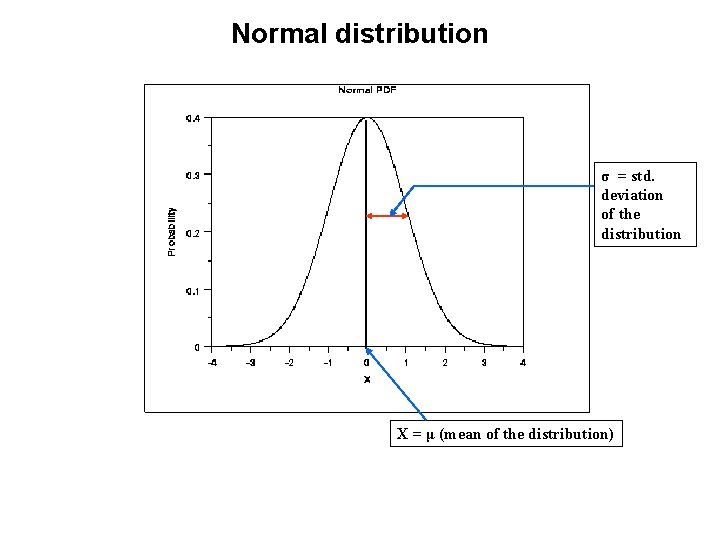
Normal distribution σ = std. deviation of the distribution X = μ (mean of the distribution)

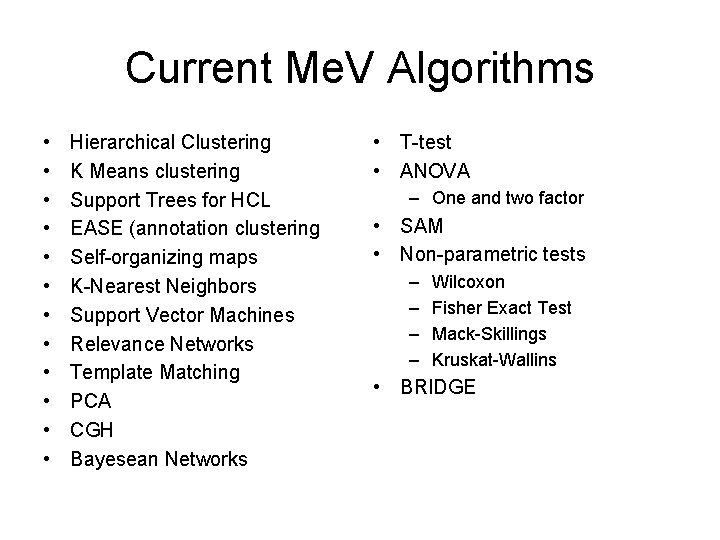
Current Me. V Algorithms • • • Hierarchical Clustering K Means clustering Support Trees for HCL EASE (annotation clustering Self-organizing maps K-Nearest Neighbors Support Vector Machines Relevance Networks Template Matching PCA CGH Bayesean Networks • T-test • ANOVA – One and two factor • SAM • Non-parametric tests – – Wilcoxon Fisher Exact Test Mack-Skillings Kruskat-Wallins • BRIDGE

Demos • • • File loaders HTA data: ANOVA Affymetrix data: SAM Non-Parametric tests CGH

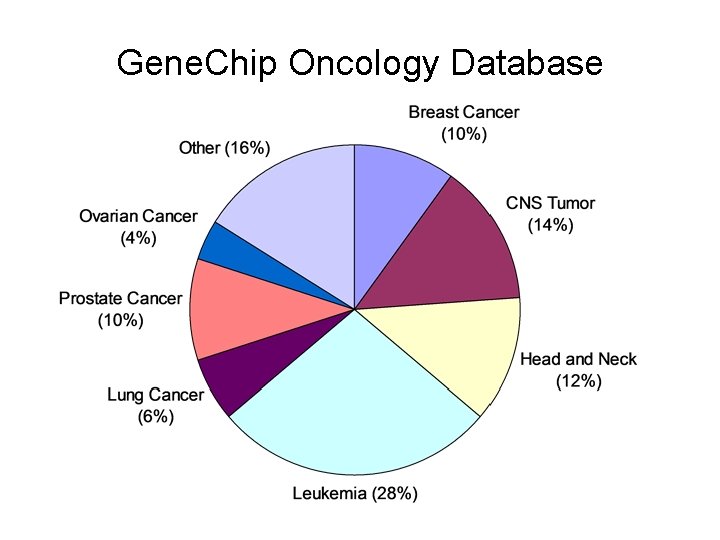
Gene. Chip Oncology Database

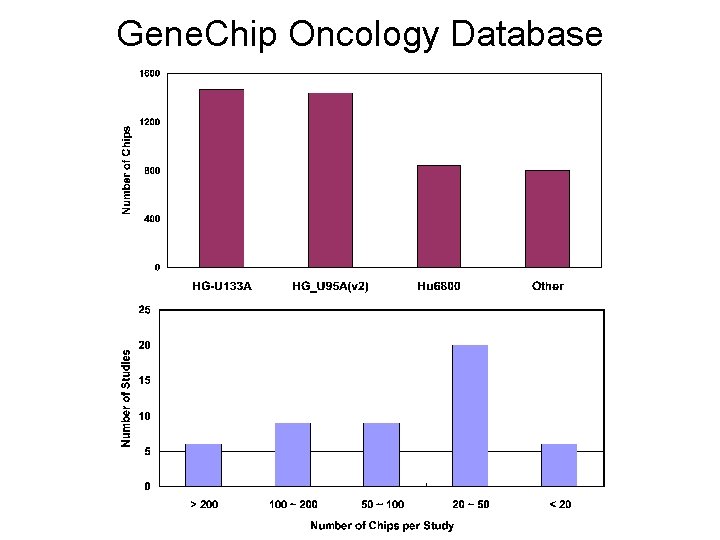
Gene. Chip Oncology Database

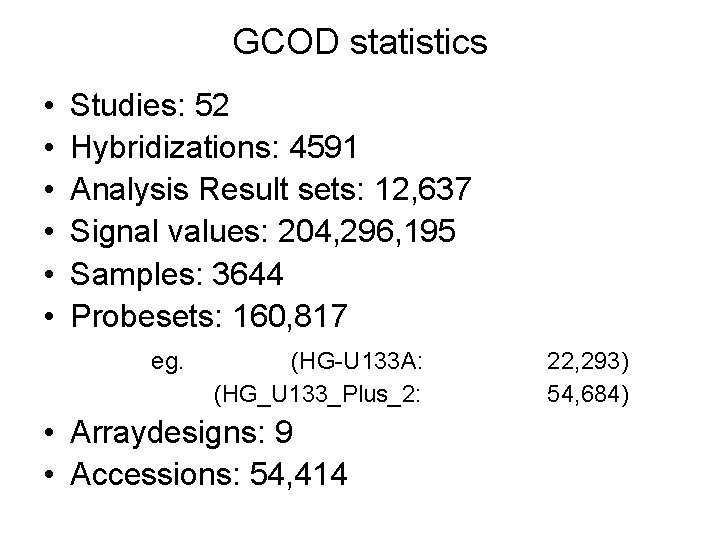
GCOD statistics • • • Studies: 52 Hybridizations: 4591 Analysis Result sets: 12, 637 Signal values: 204, 296, 195 Samples: 3644 Probesets: 160, 817 eg. (HG-U 133 A: (HG_U 133_Plus_2: • Arraydesigns: 9 • Accessions: 54, 414 22, 293) 54, 684)

Me. V Team • Eleanor Howe • Sarita Nair • Raktim Sinha • mev@tigr. org