An overview of Microarray Technology and Data Analysis










- Slides: 32

An overview of Microarray Technology and Data Analysis Basic Data Analysis

The Illumina Beadarray Technology • Highly redundant (~50 copies of a bead) • 60 mer oligos • Each array is deconvoluted using a colour coding tag system • Human, Mouse, Rat, Custom

Affymetrix Technology • Highly redundant (~25 short oligos per gene) • PM-MM oligo system valuable for cross hybe detection • Human, Mouse, E. coli, Yeast……. . • Affy and illumina arrays have been systematically compared

Spotted Arrays • • Low redundancy c. DNA and oligo Cy 5/Cy 3 dye Cost and custom

Worked Example: illumina data • Data contains 36 experiments by 47294 genes. Raw data extracted using Beadstudio. • Quality controlled in “R” package. Removed unexpressed genes using the Beadstudio Detection P-value. Leaves ~28, 000 genes. • Quantile Normalised data, and quality controlled the normalisation in ma. Corr. Plot “R” package. • Clustered using Hierarchical methods

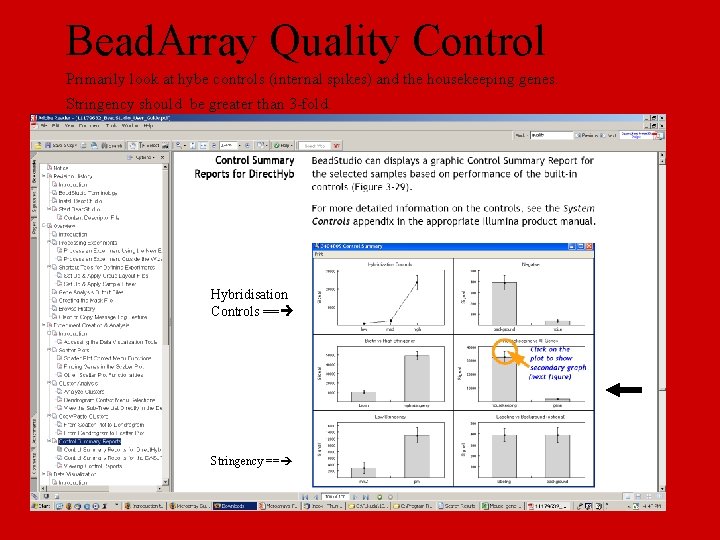
Bead. Array Quality Control Primarily look at hybe controls (internal spikes) and the housekeeping genes. Stringency should be greater than 3 -fold. Hybridisation Controls == Stringency ==

The free R-stats package A massively powerful program with hundreds of plugins BUT requires a LARGE investment to learn. Some good web resources: Bioconductor Gives you access to good free Affy analysis tools

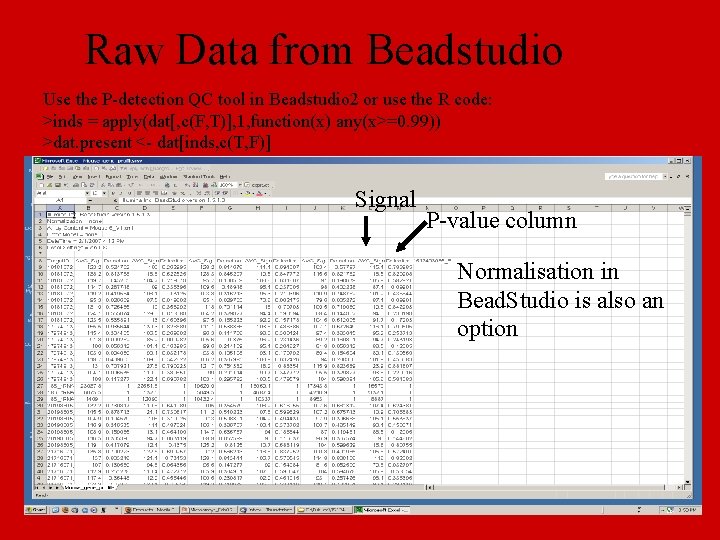
Raw Data from Beadstudio Use the P-detection QC tool in Beadstudio 2 or use the R code: >inds = apply(dat[, c(F, T)], 1, function(x) any(x>=0. 99)) >dat. present <- dat[inds, c(T, F)] Signal P-value column Normalisation in Bead. Studio is also an option

Normalisation • • • Why? Remove chip to chip variation Many different methods A) Normalisation to the mean (old school) B) Intensity-dependent normalisation – -to rank invariant genes (housekeeping) – -Quantile normalisation

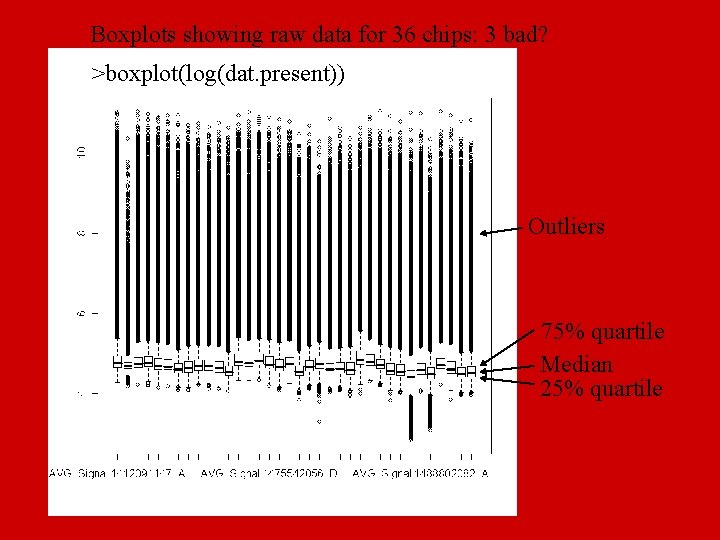
Boxplots showing raw data for 36 chips: 3 bad? >boxplot(log(dat. present)) Outliers 75% quartile Median 25% quartile

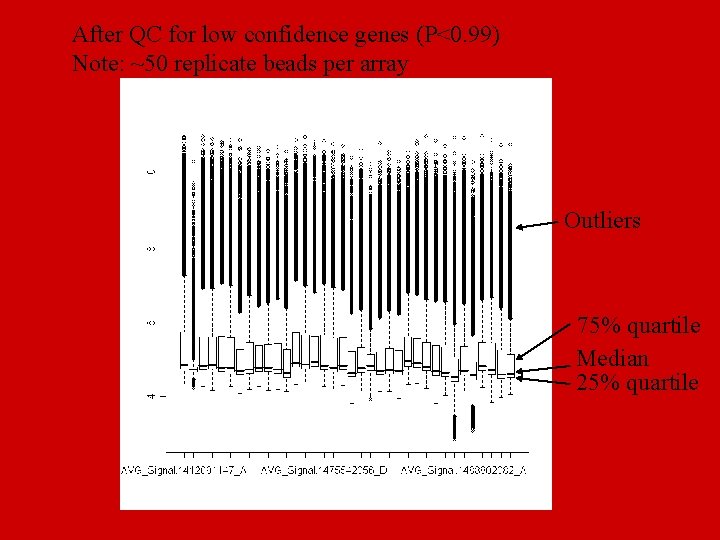
After QC for low confidence genes (P<0. 99) Note: ~50 replicate beads per array Outliers 75% quartile Median 25% quartile

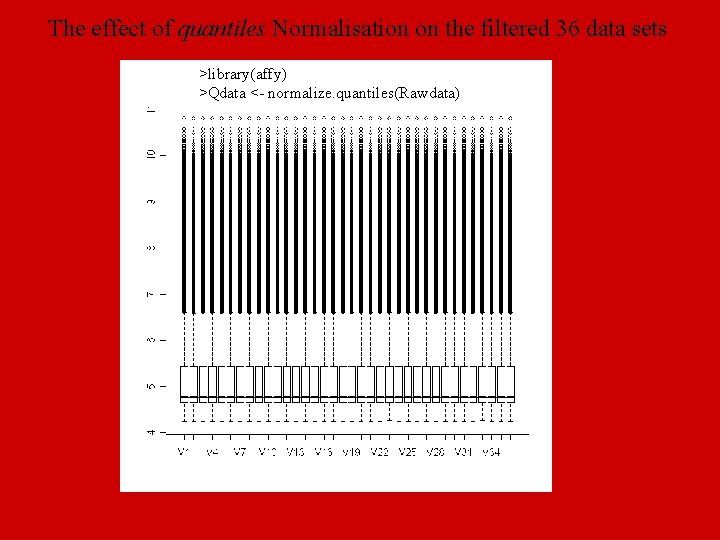
The effect of quantiles Normalisation on the filtered 36 data sets >library(affy) >Qdata <- normalize. quantiles(Rawdata)

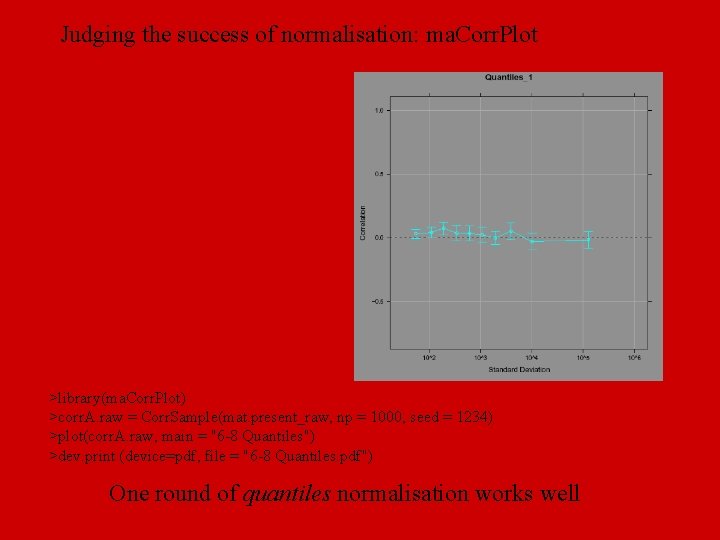
Judging the success of normalisation: ma. Corr. Plot >library(ma. Corr. Plot) >corr. A. raw = Corr. Sample(mat. present_raw, np = 1000, seed = 1234) >plot(corr. A. raw, main = "6 -8 Quantiles") >dev. print (device=pdf, file = "6 -8 Quantiles. pdf") One round of quantiles normalisation works well

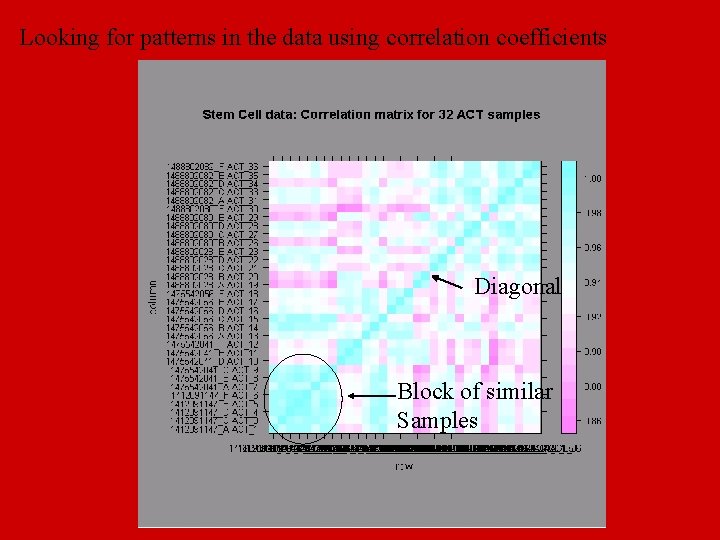
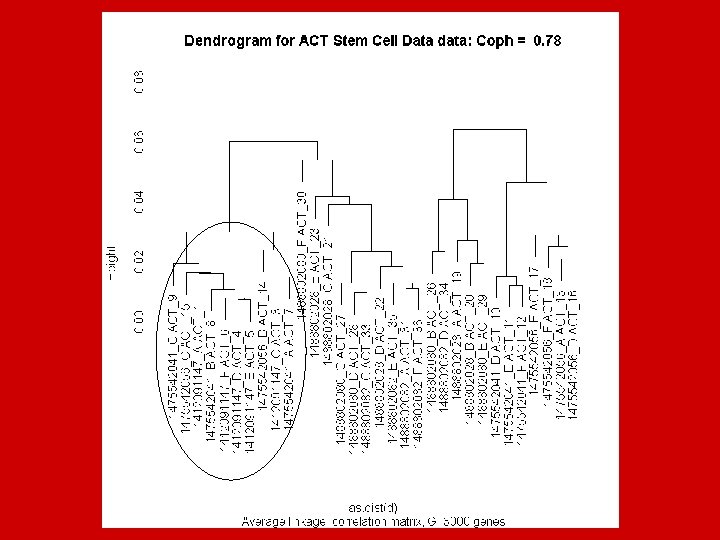
Looking for patterns in the data using correlation coefficients Diagonal Block of similar Samples


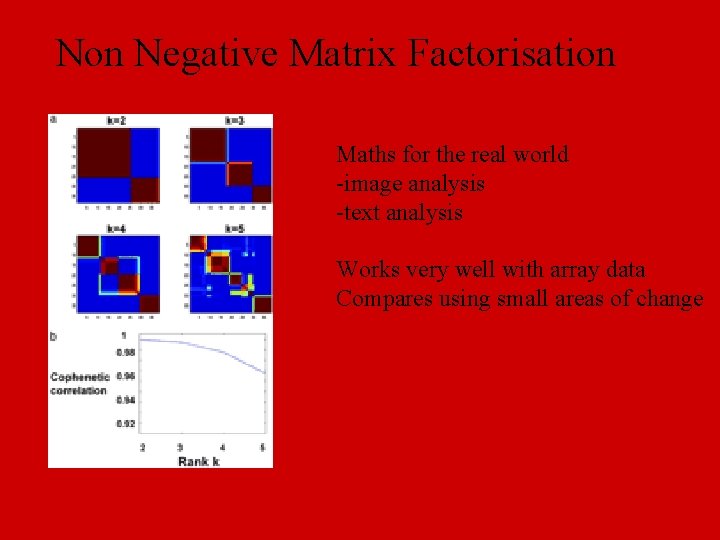
Non Negative Matrix Factorisation Maths for the real world -image analysis -text analysis Works very well with array data Compares using small areas of change

NMF: cancer classification etc Good way to visualize large data sets

Gene. Spring • Shared Resources has a copy which is available via Remote Desktop • High quality software; very carefully put together. Respected, tried and tested. • Good user friendly statistics

Core Gene. Spring functions • • • Drag and drop data table Remove low expressing genes Define replicates and groups ANOVA Expression across Pathways

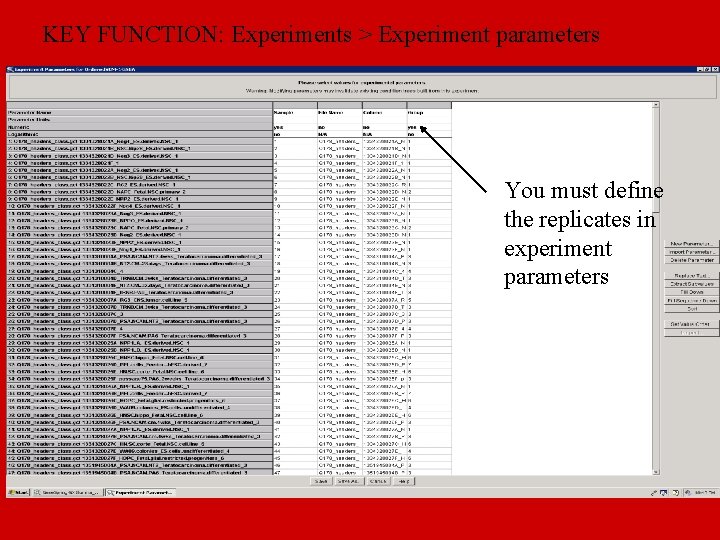
KEY FUNCTION: Experiments > Experiment parameters You must define the replicates in experiment parameters

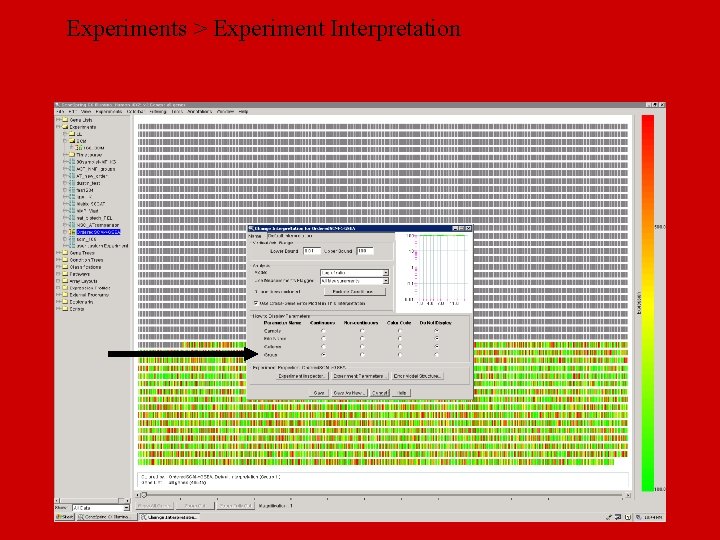
Experiments > Experiment Interpretation

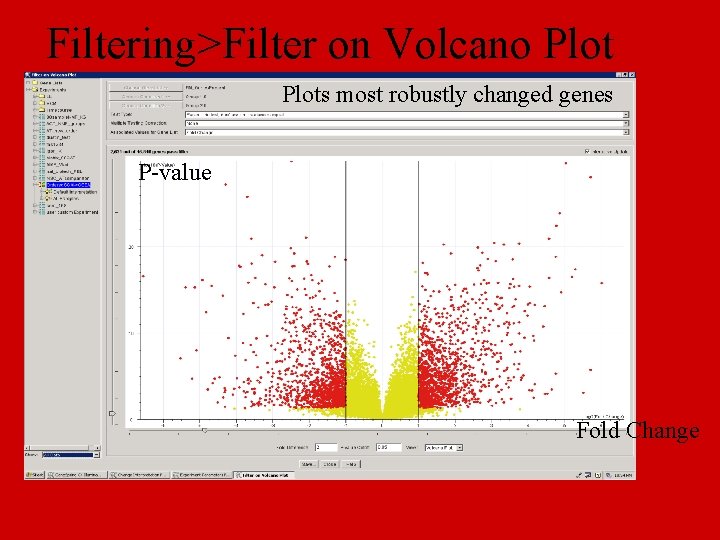
Filtering>Filter on Volcano Plots most robustly changed genes P-value Fold Change

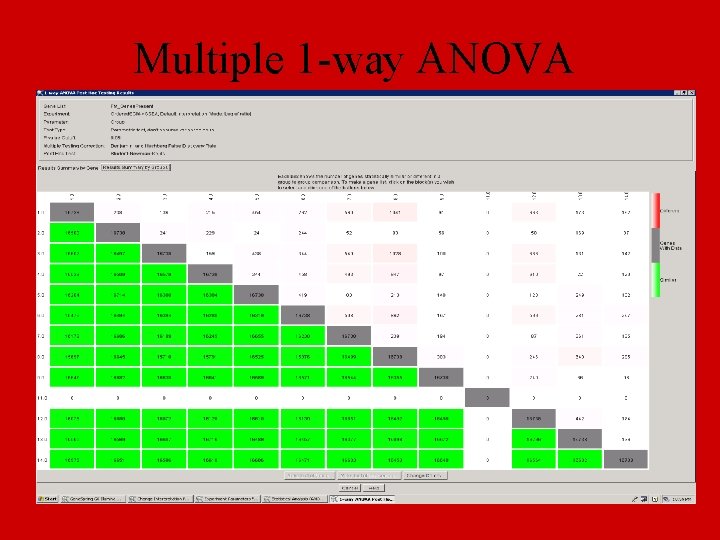
Multiple 1 -way ANOVA

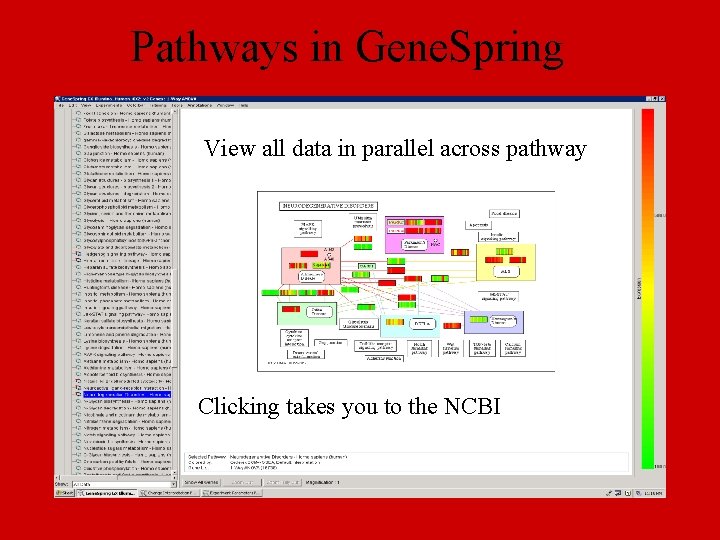
Pathways in Gene. Spring View all data in parallel across pathway Clicking takes you to the NCBI

The Free Gene. Set Enrichment Analysis (GSEA) Program • where single-gene analysis finds little similarity between two independent studies, GSEA reveals many biological pathways in common • GSEA has a database of 1, 325 biologically defined gene sets

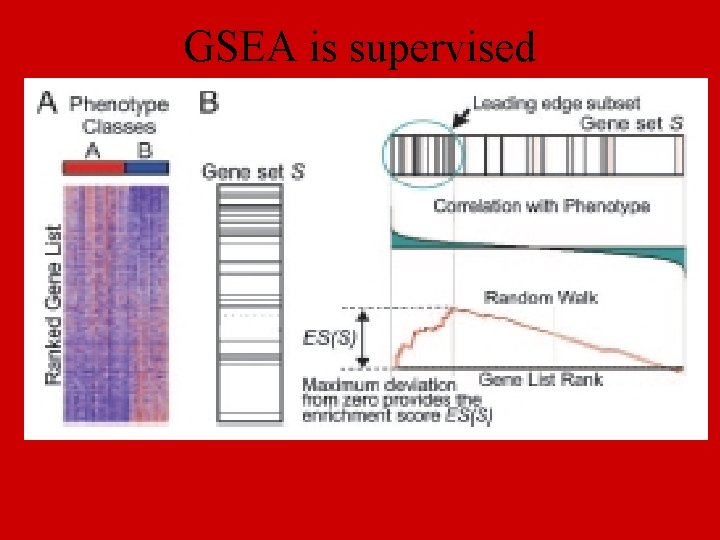
GSEA is supervised

Make *. gct and *. cls files


Monitoring Transcription Factor Regulons across cell types Network analysis

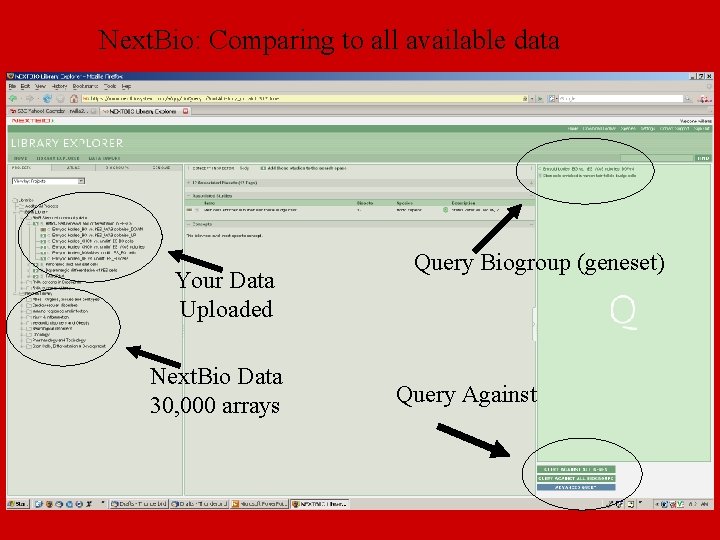
Next. Bio: Comparing to all available data Your Data Uploaded Next. Bio Data 30, 000 arrays Query Biogroup (geneset) Query Against

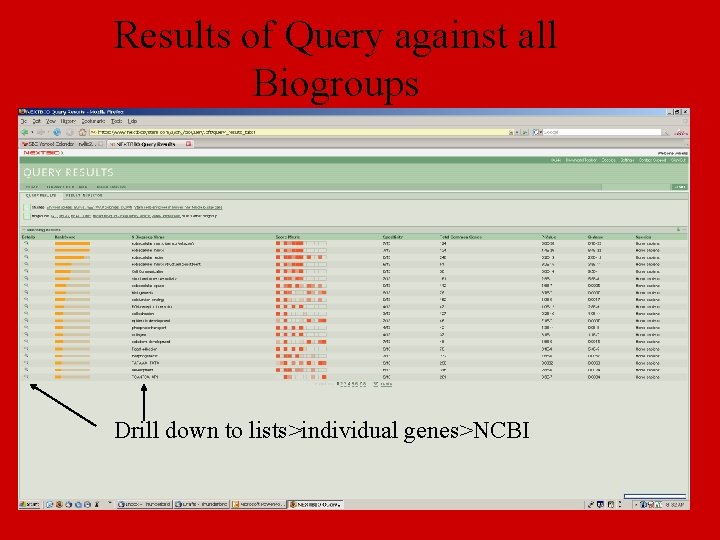
Results of Query against all Biogroups Drill down to lists>individual genes>NCBI

Dividing Biological Space