An Extensible ModelBased Mediator System with Domain Maps
An Extensible Model-Based Mediator System with Domain Maps Amarnath Gupta* Bertram Ludäscher* Maryann E. Martone+ *San Diego Supercomputer Center (SDSC) +National Center for Microscopy and Imaging Research (NCMIR) University of California, San Diego (UCSD)
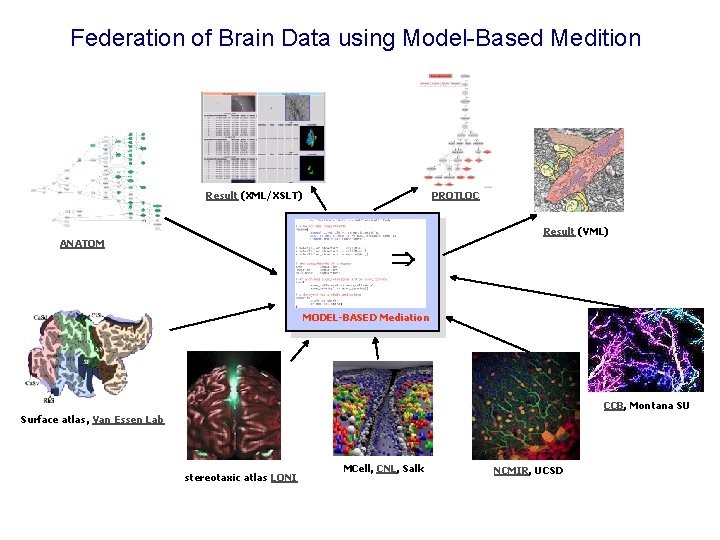
Federation of Brain Data using Model-Based Medition Result (XML/XSLT) PROTLOC Result (VML) ANATOM MODEL-BASED Mediation CCB, Montana SU Surface atlas, Van Essen Lab stereotaxic atlas LONI MCell, CNL, Salk NCMIR, UCSD
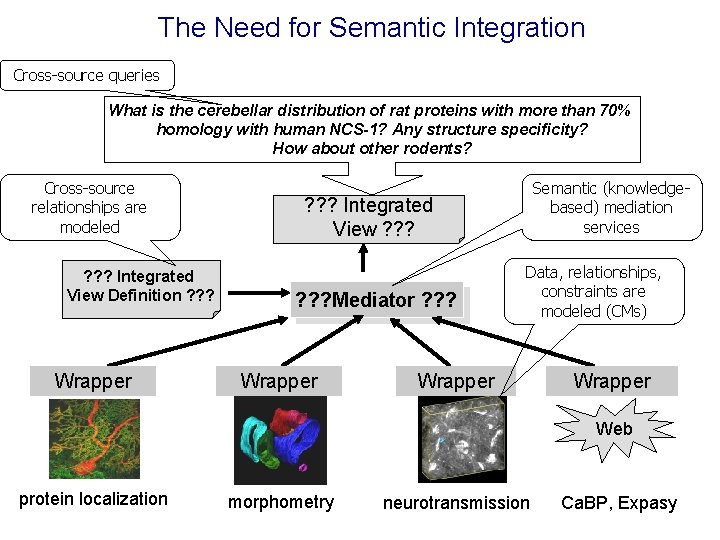
The Need for Semantic Integration Cross-source queries What is the cerebellar distribution of rat proteins with more than 70% homology with human NCS-1? Any structure specificity? How about other rodents? Cross-source relationships are modeled ? ? ? Integrated View Definition ? ? ? Wrapper Semantic (knowledgebased) mediation services ? ? ? Integrated View ? ? ? Mediator ? ? ? Wrapper Data, relationships, constraints are modeled (CMs) Wrapper Web protein localization morphometry neurotransmission Ca. BP, Expasy
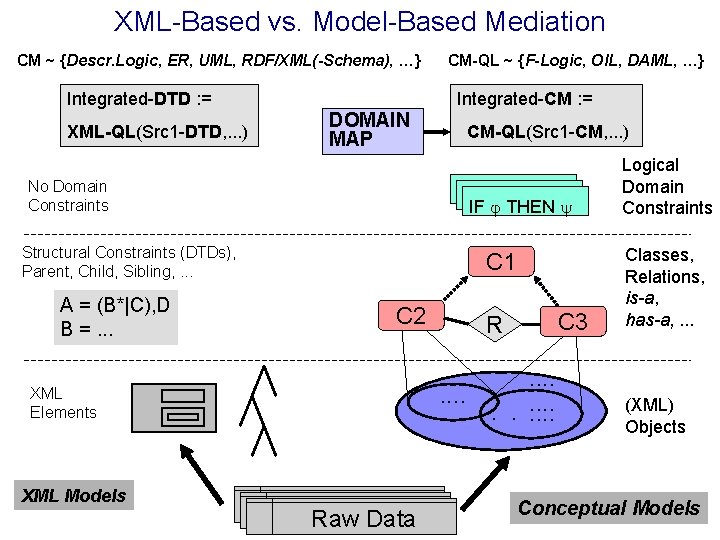
XML-Based vs. Model-Based Mediation CM ~ {Descr. Logic, ER, UML, RDF/XML(-Schema), …} Integrated-DTD : = XML-QL(Src 1 -DTD, . . . ) CM-QL ~ {F-Logic, OIL, DAML, …} Integrated-CM : = DOMAIN MAP No Domain Constraints CM-QL(Src 1 -CM, . . . ) IF THEN IF IF THEN Structural Constraints (DTDs), Parent, Child, Sibling, . . . A = (B*|C), D B =. . . C 1 C 2. . XML Elements XML Models Raw Data Raw. Data C 3 R. . . Logical Domain Constraints Classes, Relations, is-a, has-a, . . . (XML) Objects Conceptual Models
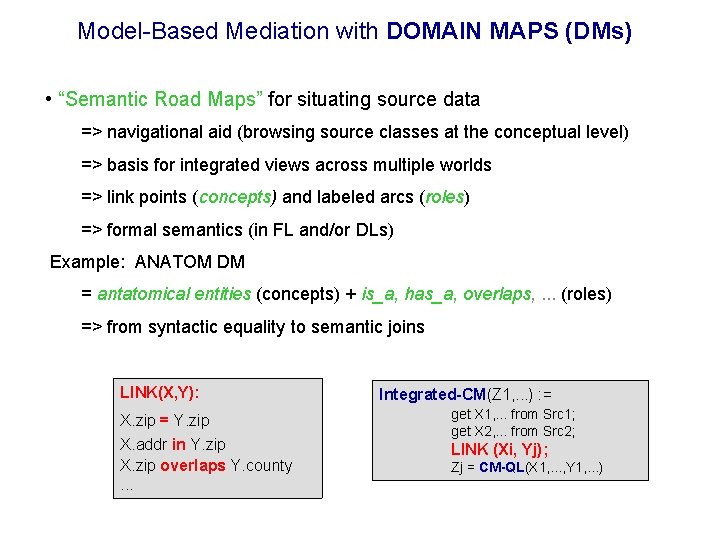
Model-Based Mediation with DOMAIN MAPS (DMs) • “Semantic Road Maps” for situating source data => navigational aid (browsing source classes at the conceptual level) => basis for integrated views across multiple worlds => link points (concepts) and labeled arcs (roles) => formal semantics (in FL and/or DLs) Example: ANATOM DM = antatomical entities (concepts) + is_a, has_a, overlaps, . . . (roles) => from syntactic equality to semantic joins LINK(X, Y): X. zip = Y. zip X. addr in Y. zip X. zip overlaps Y. county. . . Integrated-CM(Z 1, . . . ) : = get X 1, . . . from Src 1; get X 2, . . . from Src 2; LINK (Xi, Yj); Zj = CM-QL(X 1, . . . , Y 1, . . . )
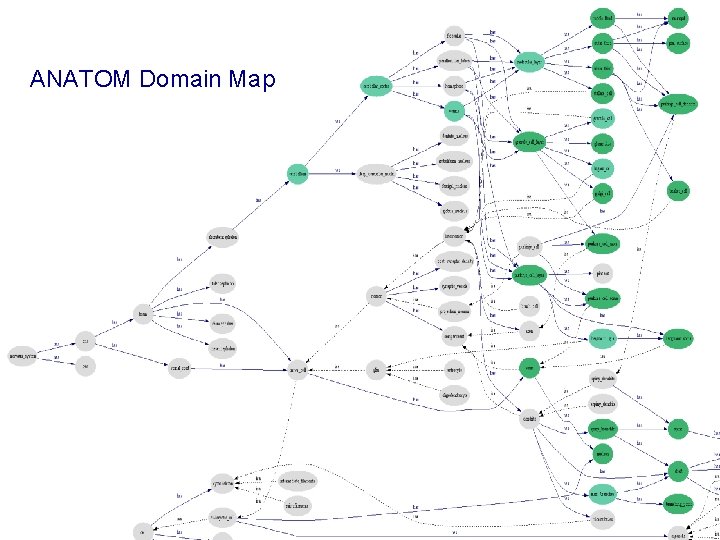
ANATOM Domain Map
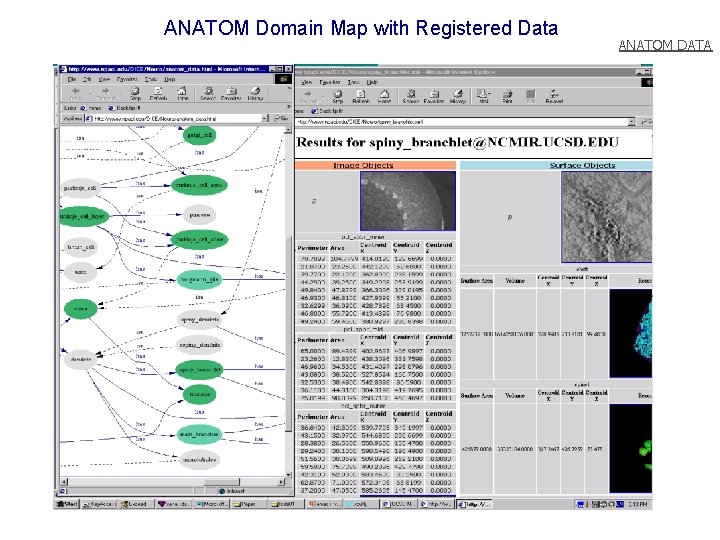
ANATOM Domain Map with Registered Data ANATOM DATA
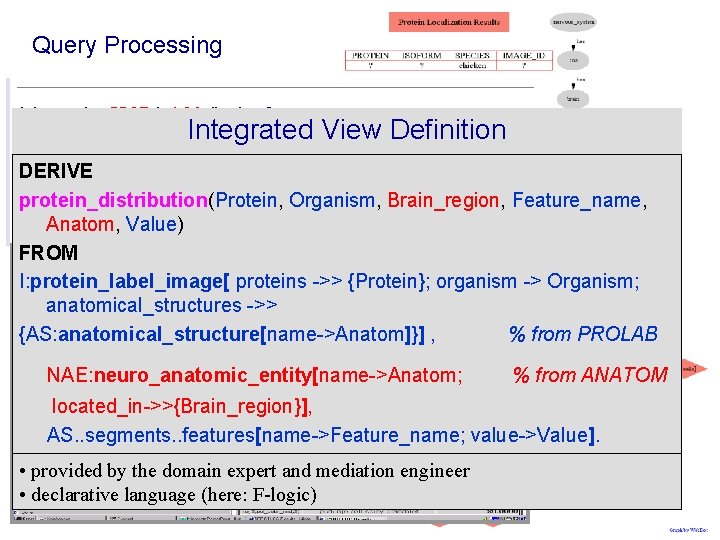
Query Processing Integrated View Definition DERIVE protein_distribution(Protein, Organism, Brain_region, Feature_name, Anatom, Value) FROM I: protein_label_image[ proteins ->> {Protein}; organism -> Organism; anatomical_structures ->> {AS: anatomical_structure[name->Anatom]}] , % from PROLAB NAE: neuro_anatomic_entity[name->Anatom; % from ANATOM located_in->>{Brain_region}], AS. . segments. . features[name->Feature_name; value->Value]. • provided by the domain expert and mediation engineer • declarative language (here: F-logic)
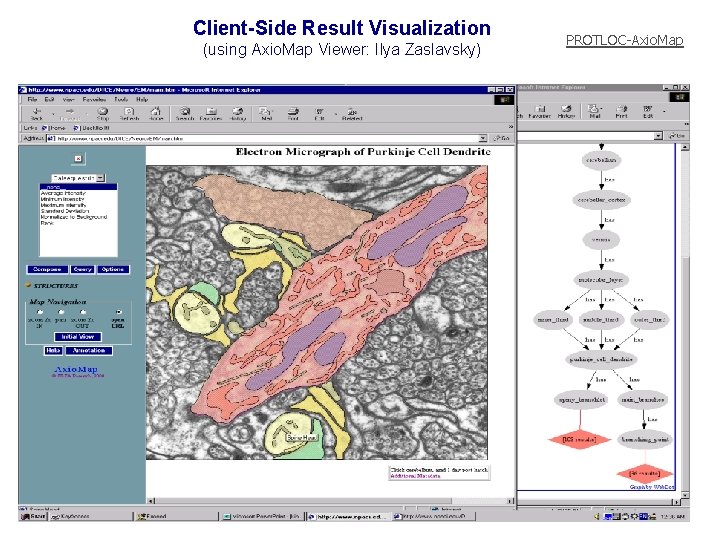
Client-Side Result Visualization (using Axio. Map Viewer: Ilya Zaslavsky) PROTLOC-Axio. Map
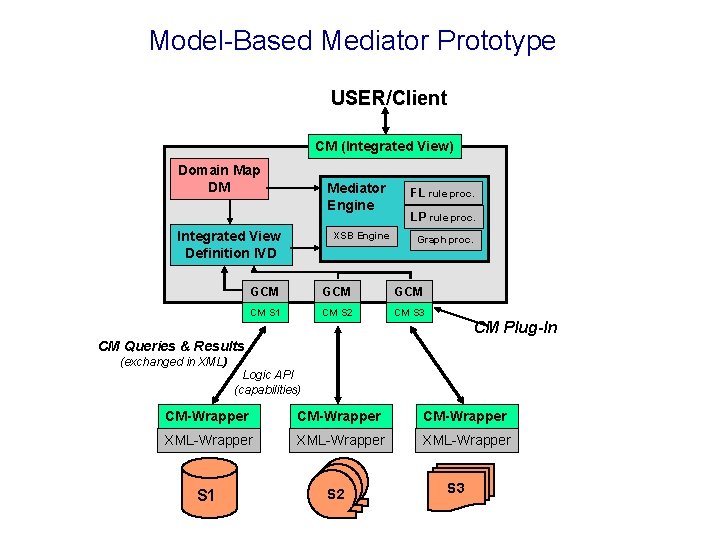
Model-Based Mediator Prototype USER/Client CM (Integrated View) Domain Map DM Mediator Engine Integrated View Definition IVD XSB Engine FL rule proc. LP rule proc. Graph proc. GCM GCM CM S 1 CM S 2 CM S 3 CM Plug-In CM Queries & Results (exchanged in XML) Logic API (capabilities) CM-Wrapper XML-Wrapper S 1 S 2 S 3
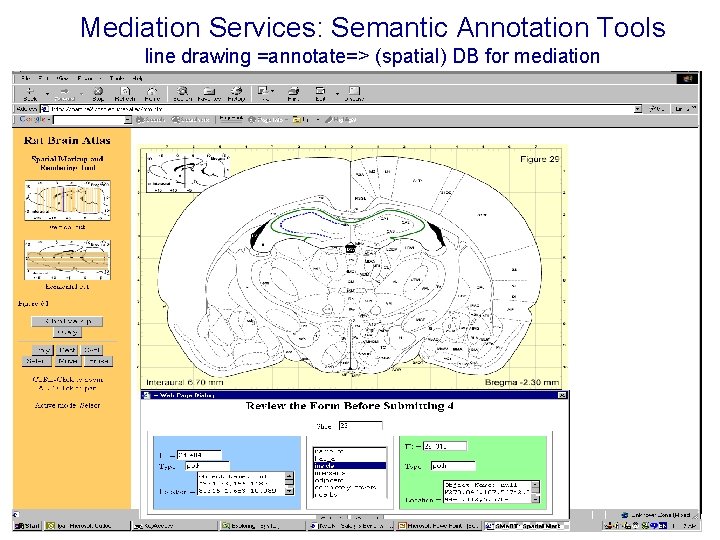
Mediation Services: Semantic Annotation Tools line drawing =annotate=> (spatial) DB for mediation
- Slides: 11