An Evolutionary Method for Training Autoencoders for Deep













- Slides: 43

An Evolutionary Method for Training Autoencoders for Deep Learning Networks MASTER ’S THESIS DEFENSE SEAN LANDE R ADVIS OR: YI SHANG University of Missouri, Department of Computer Science University of Missouri, Informatics Institute Sean Lander, Master’s Candidate

Agenda o. Overview o. Background and Related Work o. Methods o. Performance and Testing o. Results o. Conclusion and Future Work University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Agenda o. Overview o. Background and Related Work o. Methods o. Performance and Testing o. Results o. Conclusion and Future Work University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Overview Deep Learning classification/reconstruction o. Since 2006, Deep Learning Networks (DLNs) have changed the landscape of classification problems o. Strong ability to create and utilize abstract features o. Easily lends itself to GPU and distributed systems o. Does not require labeled data – VERY IMPORTANT o. Can be used for feature reduction and classification University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Overview Problem and proposed solution o. Problems with DLNs: o. Costly to train with large data sets or high feature spaces o. Local minima systemic with Artificial Neural Networks o. Hyper-parameters must be hand selected o. Proposed Solutions: o. Evolutionary based approach with local search phase o Increased chance of global minimum o Optimizes structure based on abstracted features o. Data partitions based on population size (large data only) o Reduced training time o Reduced chances of overfitting University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Agenda o. Overview o. Background and Related Work o. Methods o. Performance and Testing o. Results o. Conclusion and Future Work University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

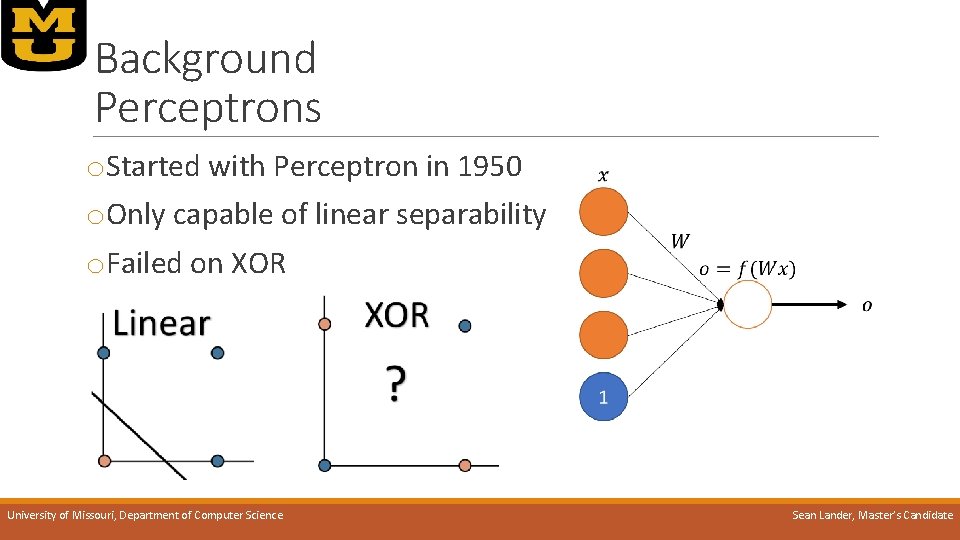
Background Perceptrons o. Started with Perceptron in 1950 o. Only capable of linear separability o. Failed on XOR University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

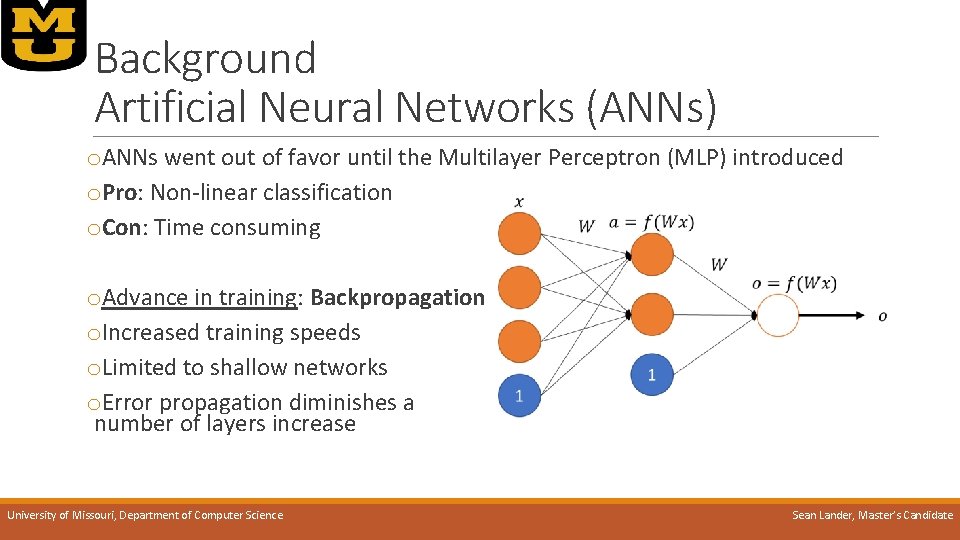
Background Artificial Neural Networks (ANNs) o. ANNs went out of favor until the Multilayer Perceptron (MLP) introduced o. Pro: Non-linear classification o. Con: Time consuming o. Advance in training: Backpropagation o. Increased training speeds o. Limited to shallow networks o. Error propagation diminishes a number of layers increase University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

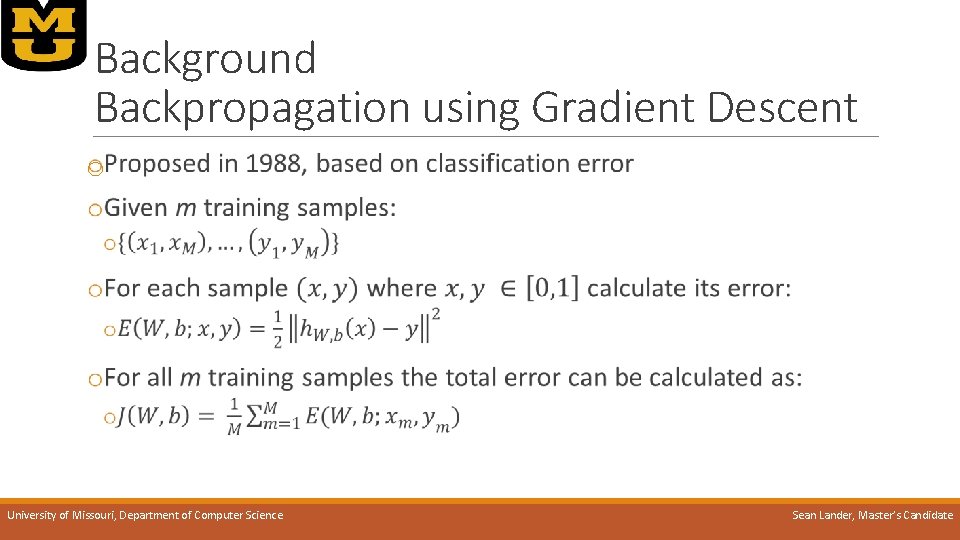
Background Backpropagation using Gradient Descent o University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

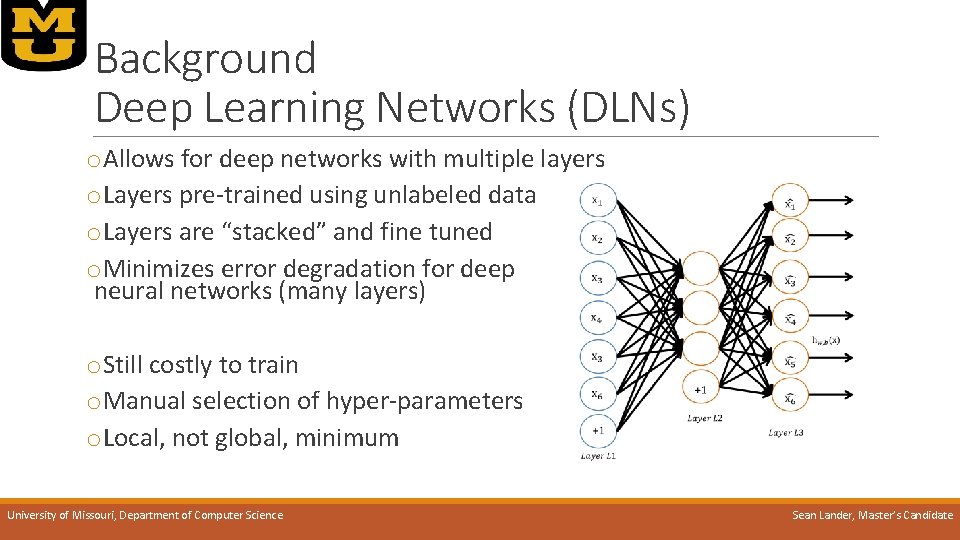
Background Deep Learning Networks (DLNs) o. Allows for deep networks with multiple layers o. Layers pre-trained using unlabeled data o. Layers are “stacked” and fine tuned o. Minimizes error degradation for deep neural networks (many layers) o. Still costly to train o. Manual selection of hyper-parameters o. Local, not global, minimum University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

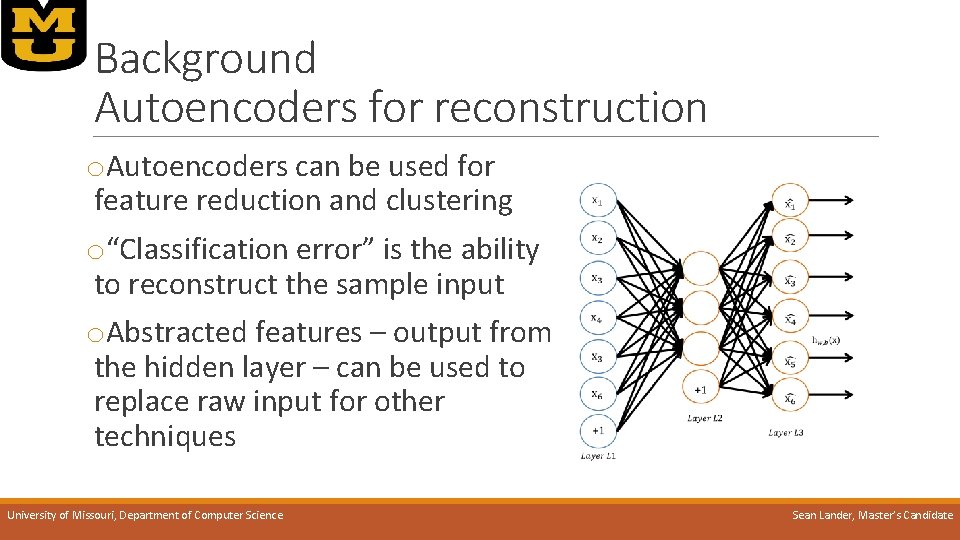
Background Autoencoders for reconstruction o. Autoencoders can be used for feature reduction and clustering o“Classification error” is the ability to reconstruct the sample input o. Abstracted features – output from the hidden layer – can be used to replace raw input for other techniques University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Related Work Evolutionary and genetic ANNs o. First use of Genetic Algorithms (GAs) in 1989 o. Two layer ANN on a small data set o. Tested multiple types of chromosomal encodings and mutation types o. Late 1990 s and early 2000 s introduced other techniques o. Multi-level mutations and mutation priority o. Addition of local search in each generation o. Inclusion of hyper-parameters as part of the mutation o. Issue of competing conventions starts to appear o Two ANNs produce the same results by sharing the same nodes but in a permuted order University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Related Work Hyper-parameter selection for DLNs o. Majority of the work explored using newer technologies and methods such as GPU and distributed (Map. Reduce) training o. Improved versions of Backpropagation, such as Conjugated Gradient or Limited Memory BFGS were tested under different conditions o. Most conclusions pointed toward manual parameter selection via trial-and-error University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Agenda o. Overview o. Background and Related Work o. Methods o. Performance and Testing o. Results o. Conclusion and Future Work University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

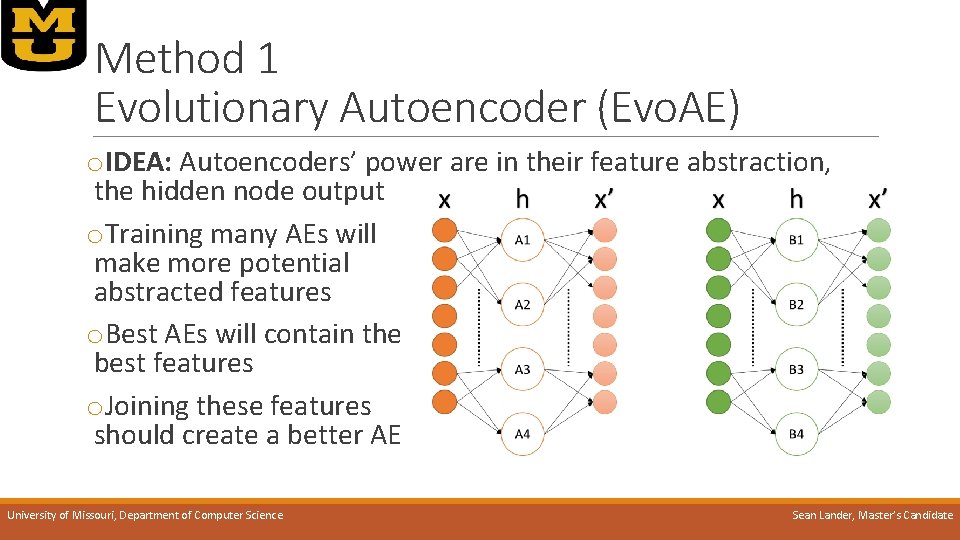
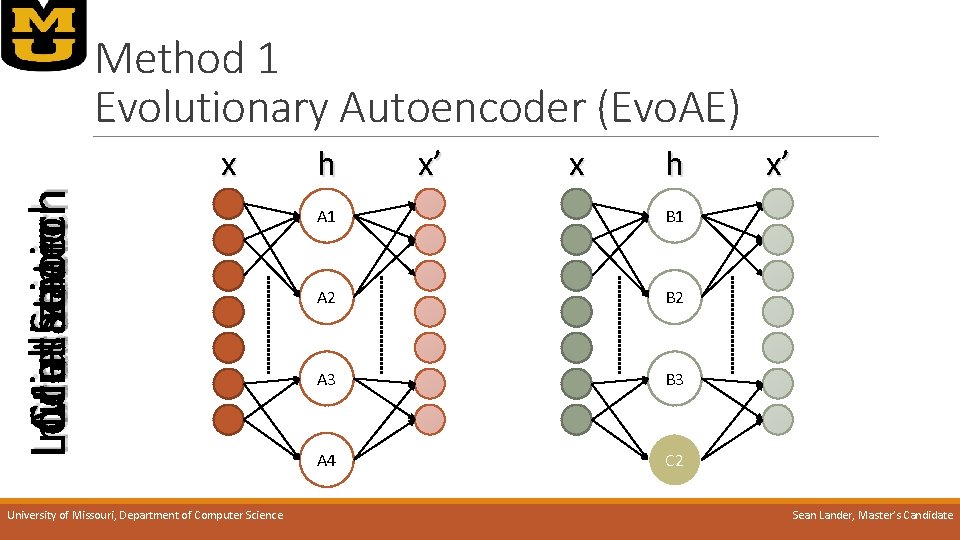
Method 1 Evolutionary Autoencoder (Evo. AE) o. IDEA: Autoencoders’ power are in their feature abstraction, the hidden node output o. Training many AEs will make more potential abstracted features o. Best AEs will contain the best features o. Joining these features should create a better AE University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Initialization Mutation Crossover Local Search Method 1 Evolutionary Autoencoder (Evo. AE) x University of Missouri, Department of Computer Science h x’ x h A 1 B 1 A 2 B 2 A 3 B 3 A 4 C 2 x’ Sean Lander, Master’s Candidate

Method 1 A Distributed learning and Mini-batches o. Training of generic Evo. AE increases in time linearly to the size of the population o. ANN training time increases drastically with data size o. To combat this, mini-batches can be used where each AE is trained against a batch and updated o. Batch size << total data University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

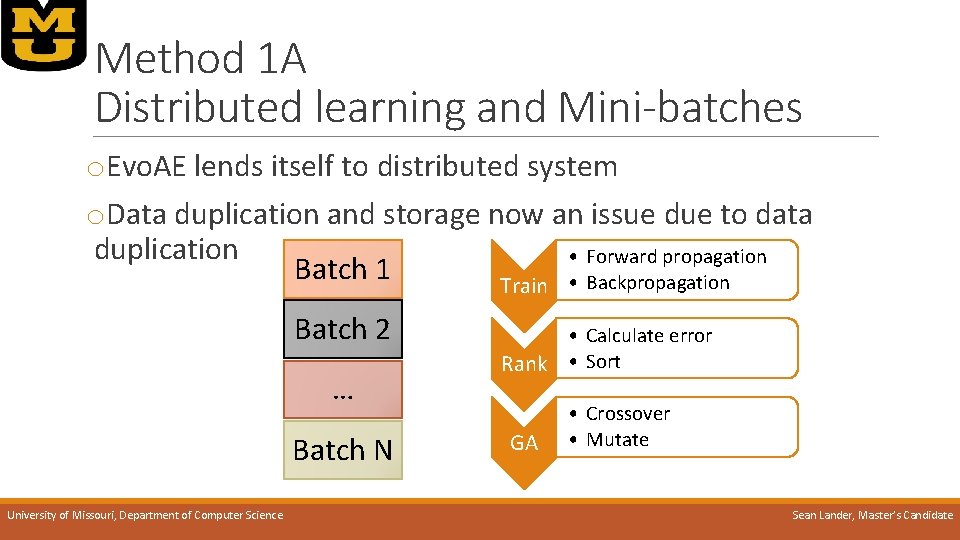
Method 1 A Distributed learning and Mini-batches o. Evo. AE lends itself to distributed system o. Data duplication and storage now an issue due to data duplication • Forward propagation Batch 1 Train • Backpropagation Batch 2 … Batch N University of Missouri, Department of Computer Science • Calculate error Rank • Sort GA • Crossover • Mutate Sean Lander, Master’s Candidate

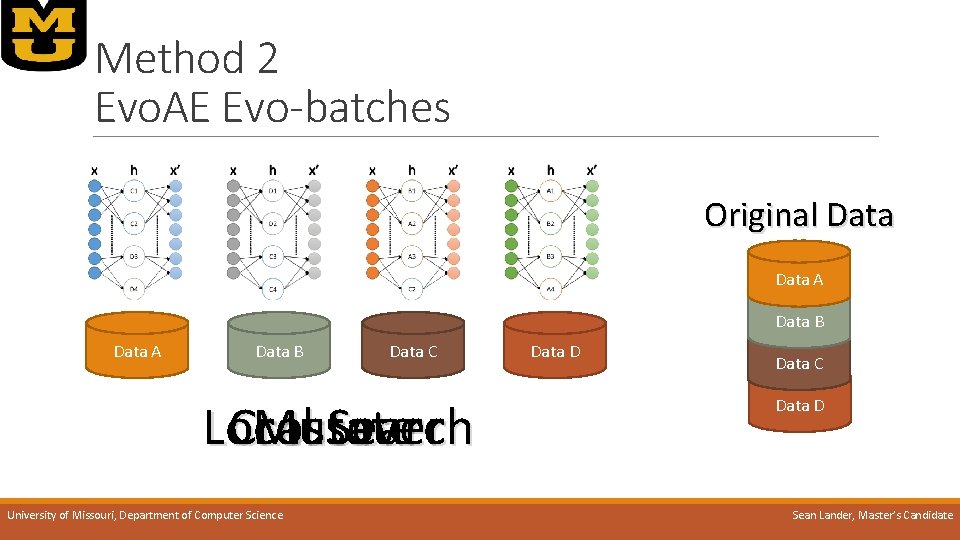
Method 2 Evo. AE Evo-batches o. IDEA: When data is large, small batches can be representative o. Prevents overfitting as nodes being trained are almost always introduced to new data o. Scales well with large amounts of data even when parallel training is not possible o. Works well on limited memory systems by increasing size of the population, thus reducing data per batch o. Quick training of large populations, equivalent to training a single autoencoder using traditional methods University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Method 2 Evo. AE Evo-batches Original Data A Data B Data C Local Crossover Mutate Search University of Missouri, Department of Computer Science Data D Data C Data D Sean Lander, Master’s Candidate

Agenda o. Overview o. Background and Related Work o. Methods o. Performance and Testing o. Results o. Conclusion and Future Work University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

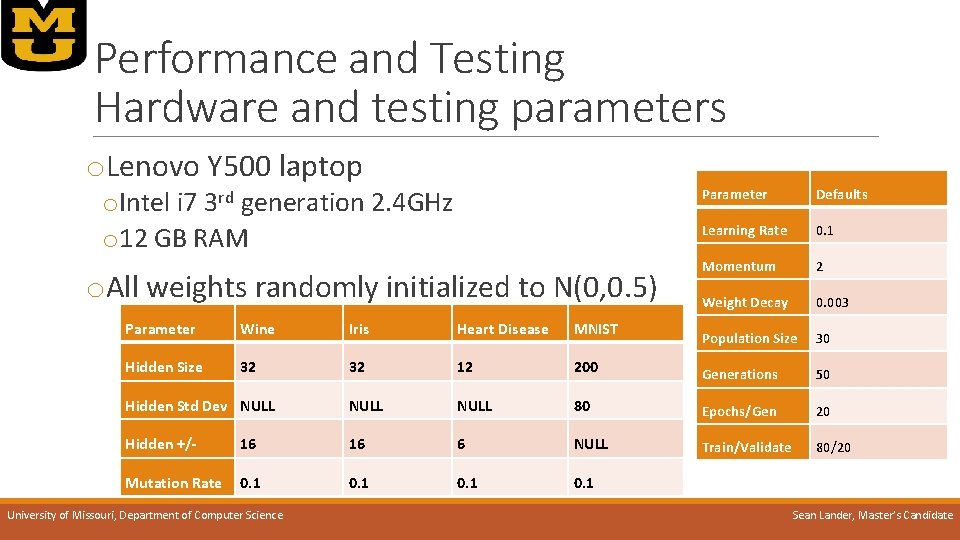
Performance and Testing Hardware and testing parameters o. Lenovo Y 500 laptop o. Intel i 7 3 rd generation 2. 4 GHz o 12 GB RAM o. All weights randomly initialized to N(0, 0. 5) Parameter Defaults Learning Rate 0. 1 Momentum 2 Weight Decay 0. 003 Population Size 30 Parameter Wine Iris Heart Disease MNIST Hidden Size 32 32 12 200 Generations 50 Hidden Std Dev NULL 80 Epochs/Gen 20 Hidden +/- 16 16 6 NULL Train/Validate 80/20 Mutation Rate 0. 1 University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

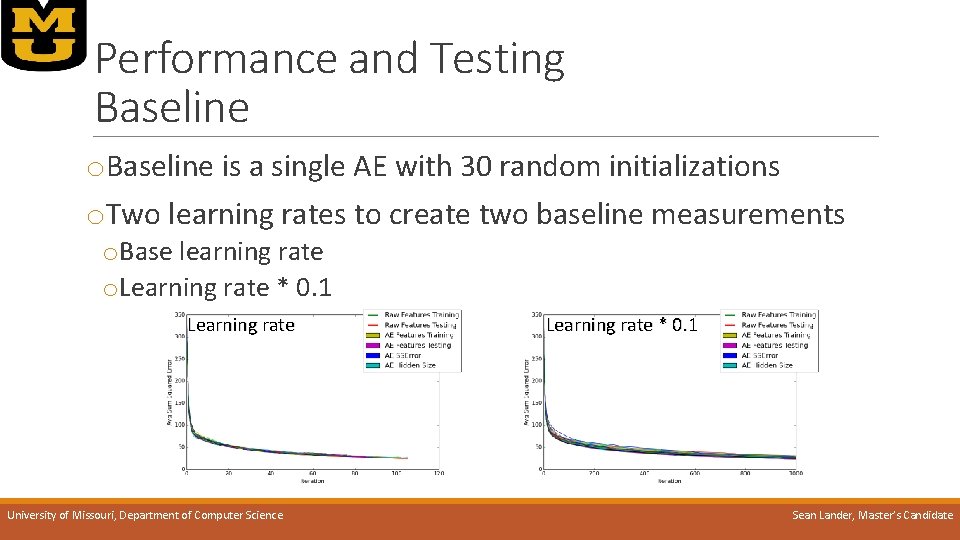
Performance and Testing Baseline o. Baseline is a single AE with 30 random initializations o. Two learning rates to create two baseline measurements o. Base learning rate o. Learning rate * 0. 1 Learning rate University of Missouri, Department of Computer Science Learning rate * 0. 1 Sean Lander, Master’s Candidate

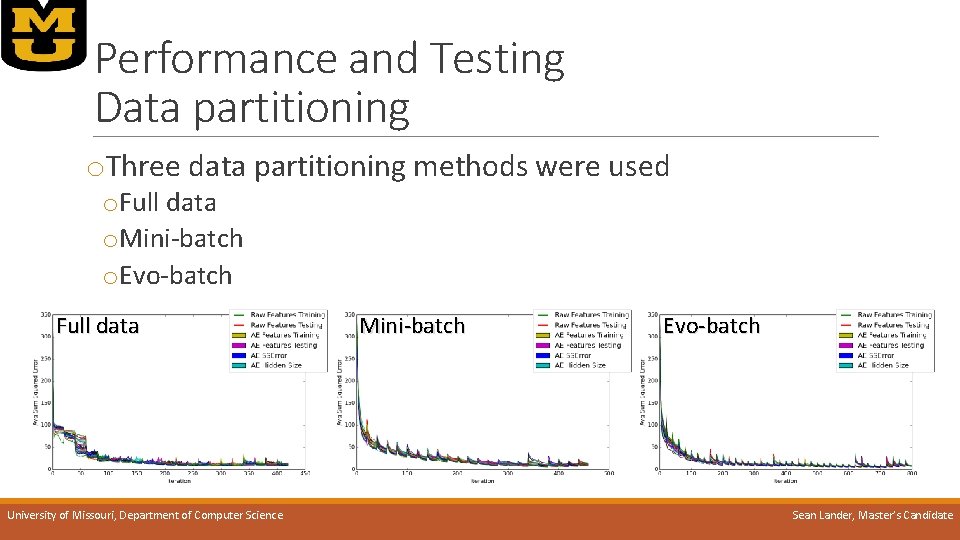
Performance and Testing Data partitioning o. Three data partitioning methods were used o. Full data o. Mini-batch o. Evo-batch Full data University of Missouri, Department of Computer Science Mini-batch Evo-batch Sean Lander, Master’s Candidate

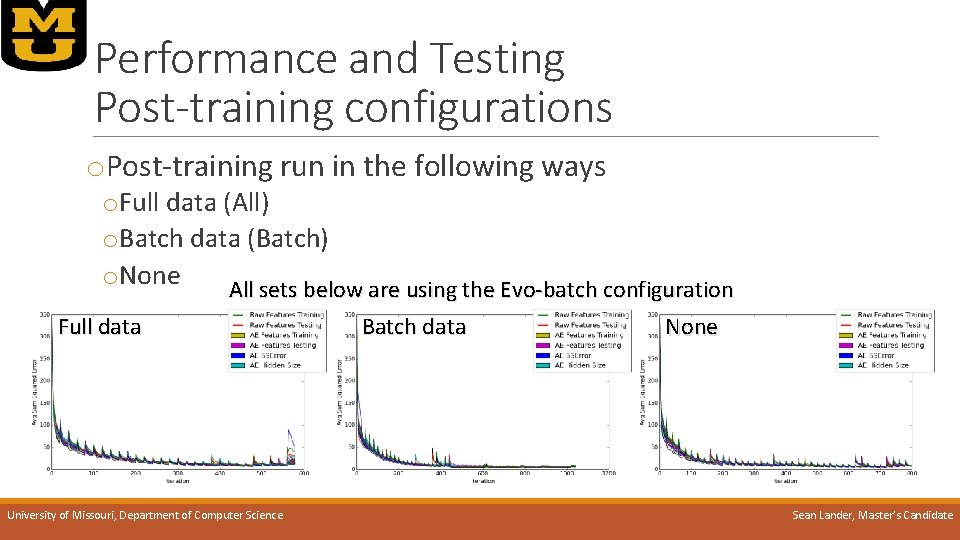
Performance and Testing Post-training configurations o. Post-training run in the following ways o. Full data (All) o. Batch data (Batch) o. None All sets below are using the Evo-batch configuration Full data University of Missouri, Department of Computer Science Batch data None Sean Lander, Master’s Candidate

Agenda o. Overview o. Background and Related Work o. Methods o. Performance and Testing o. Results o. Conclusion and Future Work University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

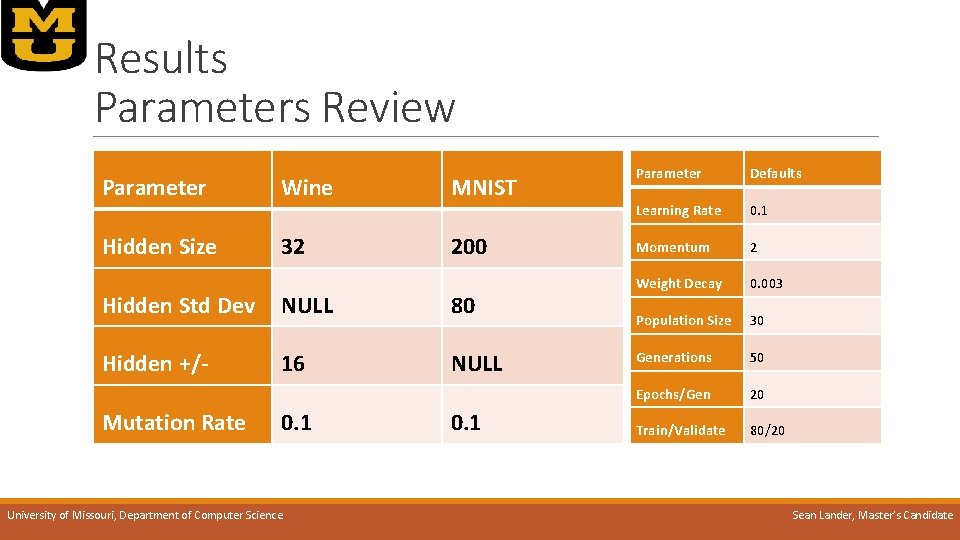
Results Parameters Review Parameter Wine MNIST Hidden Size 32 200 Hidden Std Dev NULL 80 Hidden +/- 16 NULL Mutation Rate 0. 1 University of Missouri, Department of Computer Science 0. 1 Parameter Defaults Learning Rate 0. 1 Momentum 2 Weight Decay 0. 003 Population Size 30 Generations 50 Epochs/Gen 20 Train/Validate 80/20 Sean Lander, Master’s Candidate

Results Datasets o. UCI wine dataset o 178 samples o 13 features o 3 classes o. Reduced MNIST dataset o 6000/1000 and 24 k/6 k training/testing samples o 784 features o 10 classes (0 -9) University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

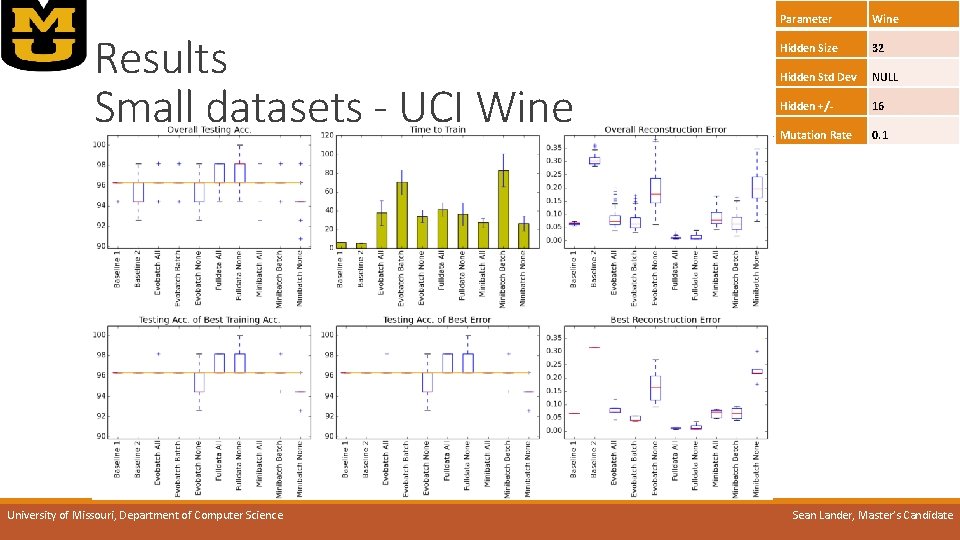
Results Small datasets - UCI Wine University of Missouri, Department of Computer Science Parameter Wine Hidden Size 32 Hidden Std Dev NULL Hidden +/- 16 Mutation Rate 0. 1 Sean Lander, Master’s Candidate

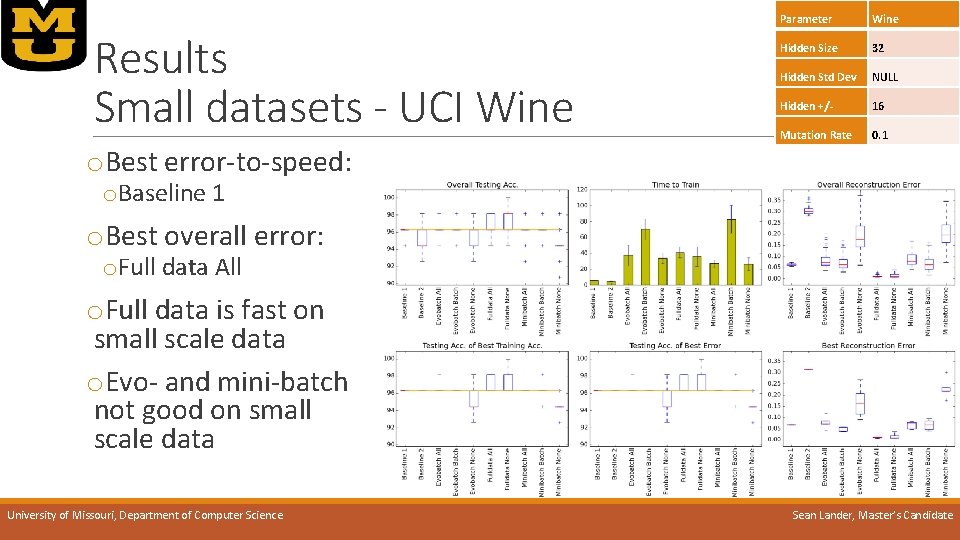
Results Small datasets - UCI Wine Parameter Wine Hidden Size 32 Hidden Std Dev NULL Hidden +/- 16 Mutation Rate 0. 1 o. Best error-to-speed: o. Baseline 1 o. Best overall error: o. Full data All o. Full data is fast on small scale data o. Evo- and mini-batch not good on small scale data University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

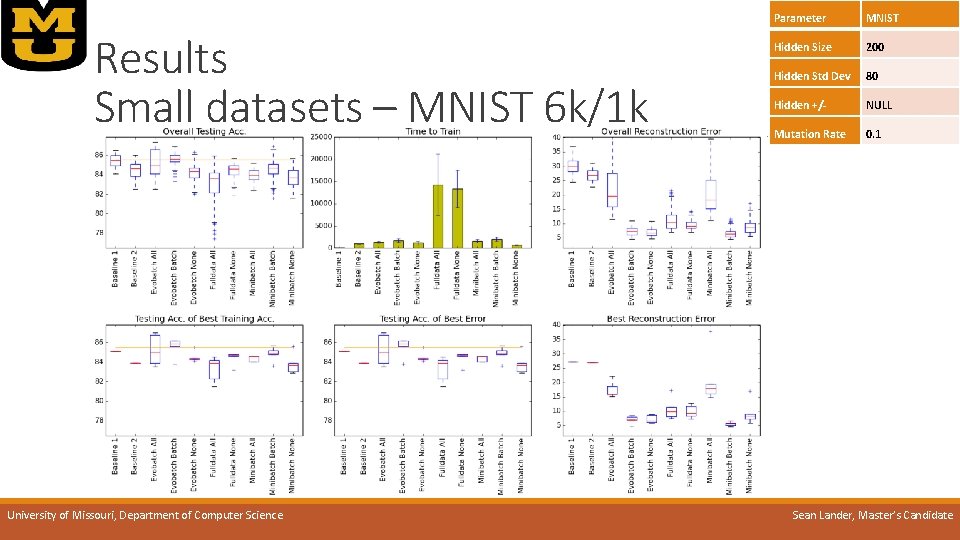
Results Small datasets – MNIST 6 k/1 k University of Missouri, Department of Computer Science Parameter MNIST Hidden Size 200 Hidden Std Dev 80 Hidden +/- NULL Mutation Rate 0. 1 Sean Lander, Master’s Candidate

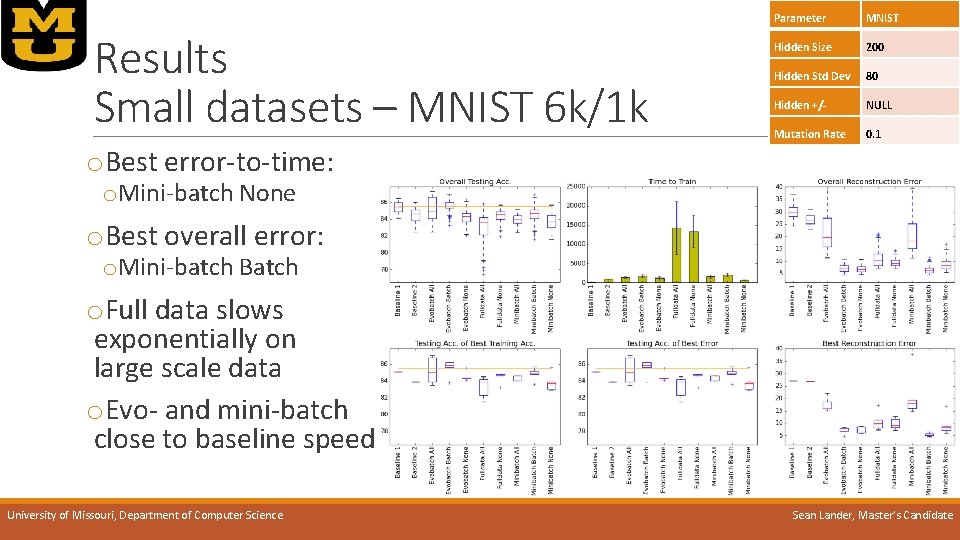
Results Small datasets – MNIST 6 k/1 k Parameter MNIST Hidden Size 200 Hidden Std Dev 80 Hidden +/- NULL Mutation Rate 0. 1 o. Best error-to-time: o. Mini-batch None o. Best overall error: o. Mini-batch Batch o. Full data slows exponentially on large scale data o. Evo- and mini-batch close to baseline speed University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

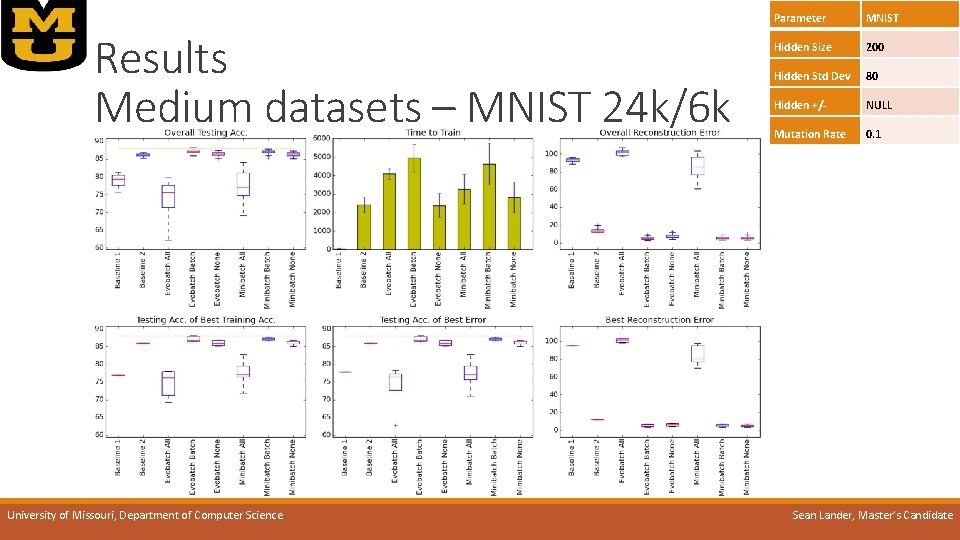
Results Medium datasets – MNIST 24 k/6 k University of Missouri, Department of Computer Science Parameter MNIST Hidden Size 200 Hidden Std Dev 80 Hidden +/- NULL Mutation Rate 0. 1 Sean Lander, Master’s Candidate

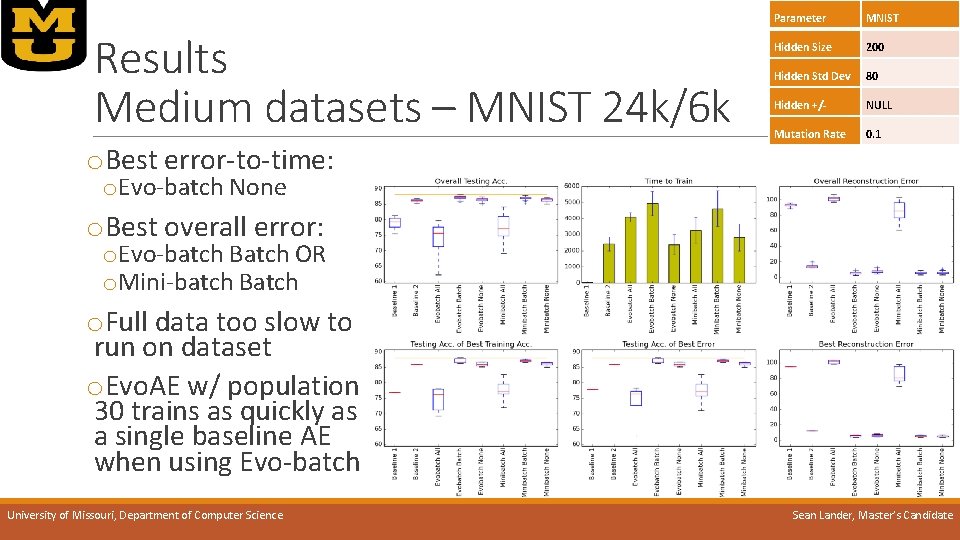
Results Medium datasets – MNIST 24 k/6 k o. Best error-to-time: Parameter MNIST Hidden Size 200 Hidden Std Dev 80 Hidden +/- NULL Mutation Rate 0. 1 o. Evo-batch None o. Best overall error: o. Evo-batch Batch OR o. Mini-batch Batch o. Full data too slow to run on dataset o. Evo. AE w/ population 30 trains as quickly as a single baseline AE when using Evo-batch University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Agenda o. Overview o. Background and Related Work o. Methods o. Performance and Testing o. Results o. Conclusion and Future Work University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Conclusions Good for large problems o. Traditional methods are still preferred choice for small problems and toy problems o. Evo. AE with Evo-batch produces effective and efficient feature reduction given a large volume of data o. Evo. AE is robust against poorly-chosen hyper-parameters, specifically learning rate University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Future Work o. Immediate goals: o. Transition to distributed system, Map. Reduce based or otherwise o. Harness GPU technology for increased speeds (~50% in some cases) o. Long term goals: o. Open the system for use by novices and non-programmers o. Make the system easy to use and transparent to the user for both modification and training purposes University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Thank you University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

Background Backpropagation with weight decay o University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

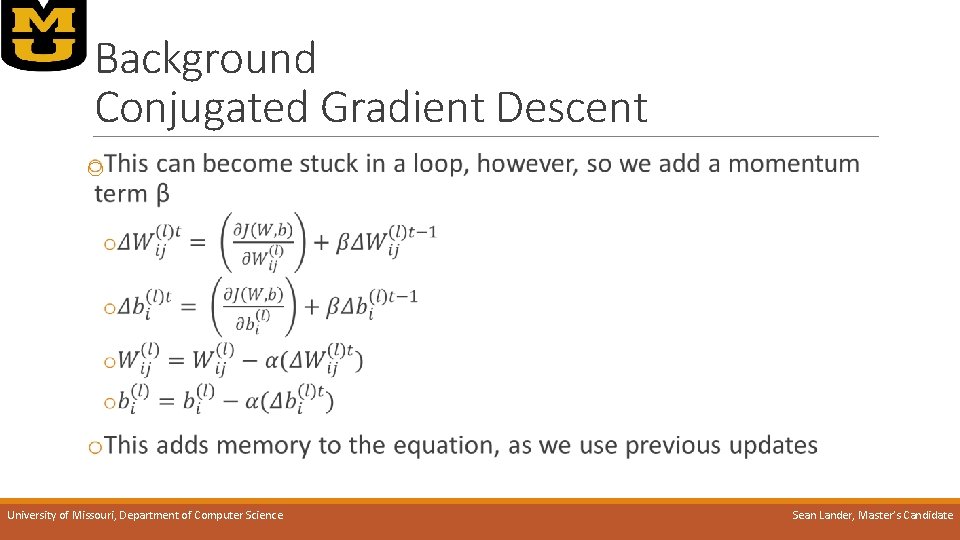
Background Conjugated Gradient Descent o University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

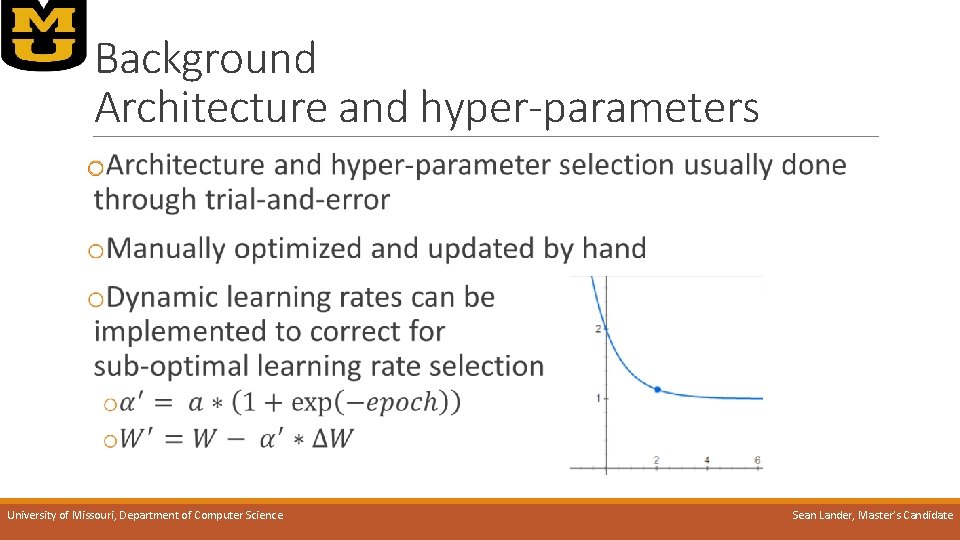
Background Architecture and hyper-parameters o University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

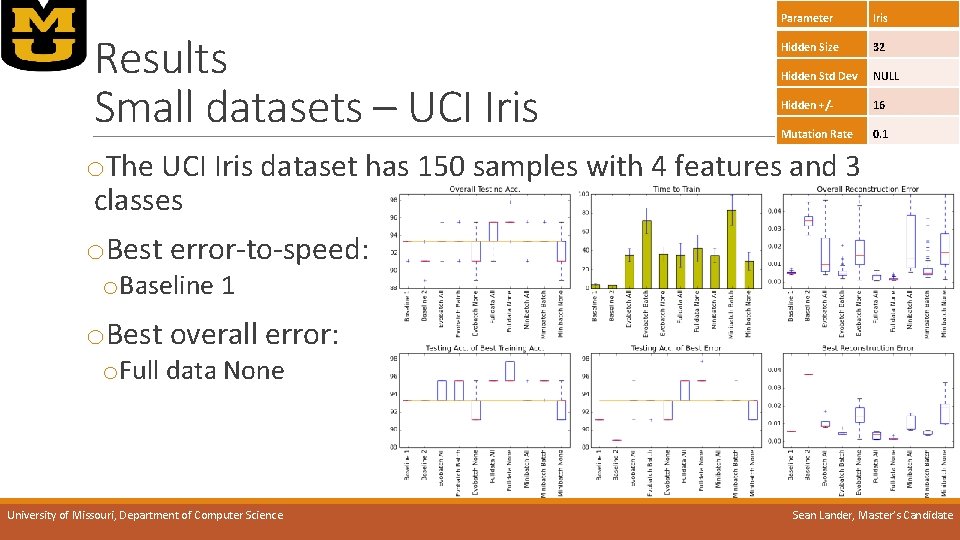
Results Small datasets – UCI Iris Parameter Iris Hidden Size 32 Hidden Std Dev NULL Hidden +/- 16 Mutation Rate 0. 1 o. The UCI Iris dataset has 150 samples with 4 features and 3 classes o. Best error-to-speed: o. Baseline 1 o. Best overall error: o. Full data None University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate

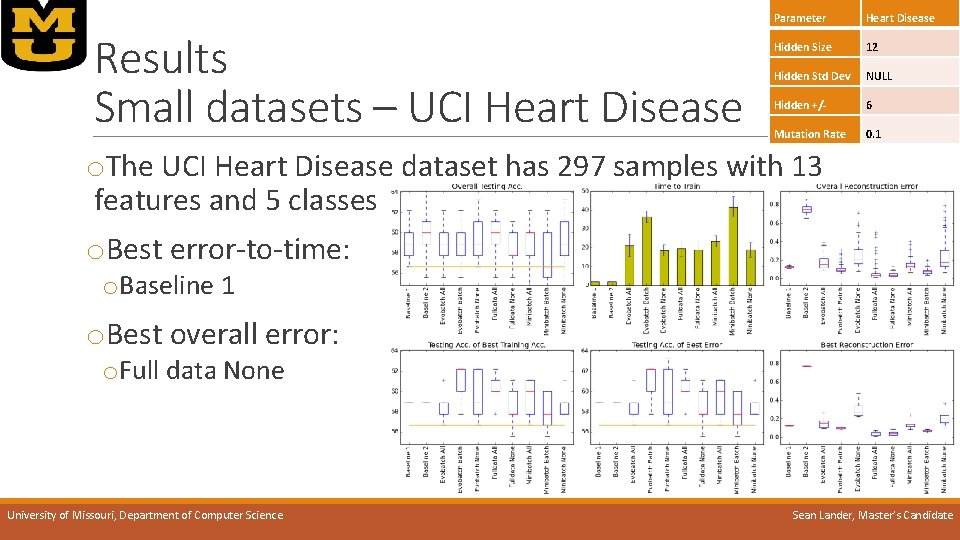
Results Small datasets – UCI Heart Disease Parameter Heart Disease Hidden Size 12 Hidden Std Dev NULL Hidden +/- 6 Mutation Rate 0. 1 o. The UCI Heart Disease dataset has 297 samples with 13 features and 5 classes o. Best error-to-time: o. Baseline 1 o. Best overall error: o. Full data None University of Missouri, Department of Computer Science Sean Lander, Master’s Candidate