An automated guidelinesbased approach for variants pathogenicity assessment

An automated guidelinesbased approach for variants pathogenicity assessment Data and Informatics-based Intervention S 28 Giovanna Nicora University of Pavia (Italy)

Disclosure I and my spouse/partner have no relevant relationships with commercial interests to disclose. AMIA 2018 | amia. org 2
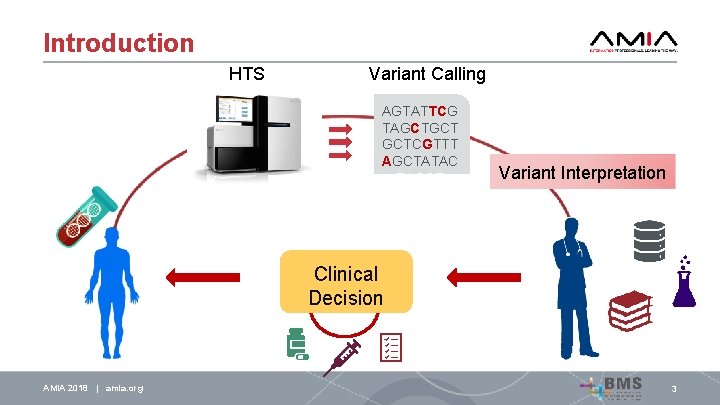
Introduction HTS Variant Calling AGTATTCG TAGCTGCT GCTCGTTT AGCTATAC TAGCT Variant Interpretation Clinical Decision AMIA 2018 | amia. org 3
Genomic Variant Interpretation AMIA 2018 | amia. org 4
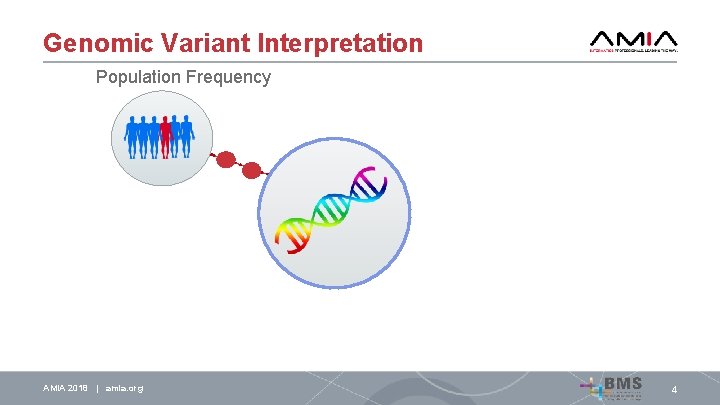
Genomic Variant Interpretation Population Frequency AMIA 2018 | amia. org 4

Genomic Variant Interpretation Population Frequency AMIA 2018 | amia. org Literature Review 4
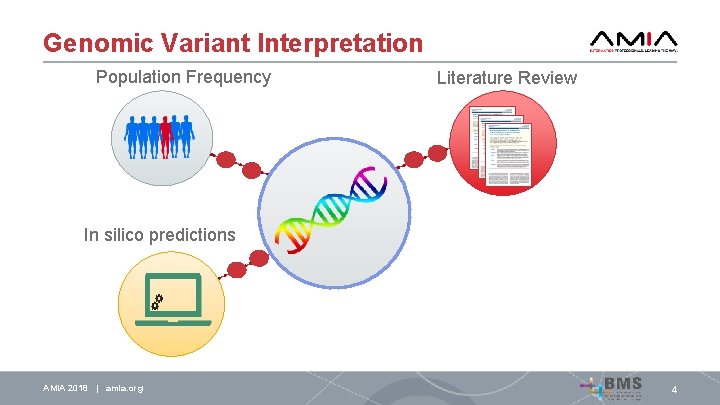
Genomic Variant Interpretation Population Frequency Literature Review In silico predictions AMIA 2018 | amia. org 4
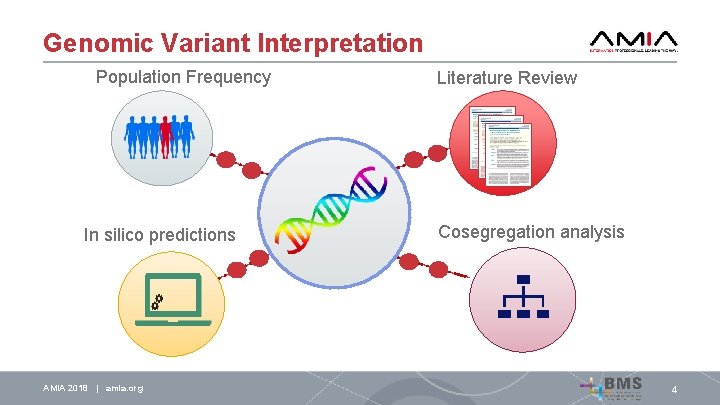
Genomic Variant Interpretation Population Frequency In silico predictions AMIA 2018 | amia. org Literature Review Cosegregation analysis 4

ACMG/AMP Standard Guidelines ü Germline variants ü Mendelian diseases AMIA 2018 | amia. org 5
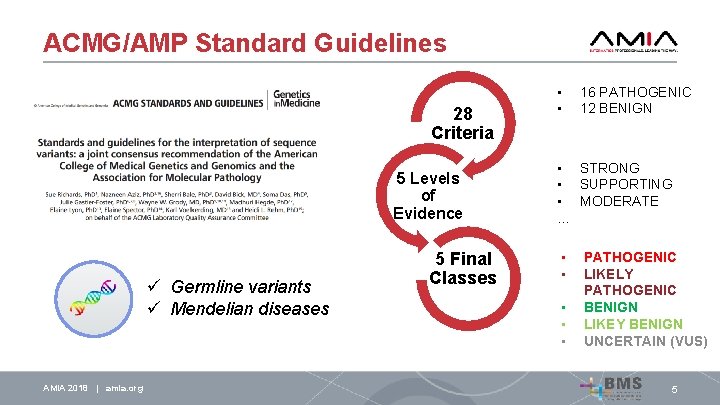
ACMG/AMP Standard Guidelines 28 Criteria 5 Levels of Evidence ü Germline variants ü Mendelian diseases 5 Final Classes • • 16 PATHOGENIC 12 BENIGN • STRONG • SUPPORTING • MODERATE … • • • PATHOGENIC LIKELY PATHOGENIC BENIGN LIKEY BENIGN UNCERTAIN (VUS) AMIA 2018 | amia. org 5

ACMG/AMP: Problems Actual application of ACMG/AMP guidelines in clinical routine is limited by: Complexity AMIA 2018 | amia. org 6

ACMG/AMP: Problems Actual application of ACMG/AMP guidelines in clinical routine is limited by: Complexity AMIA 2018 | amia. org Flexibility 6
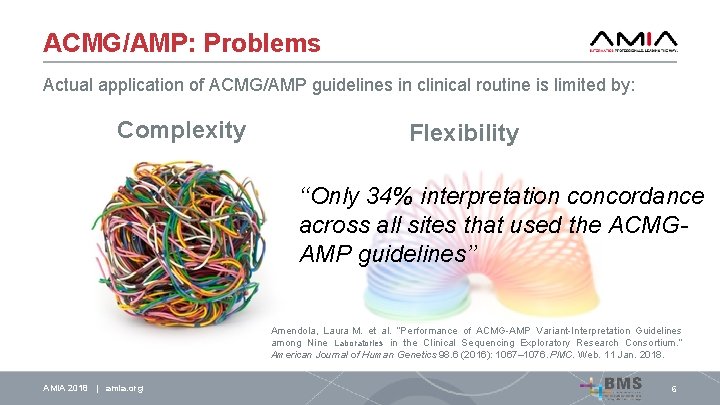
ACMG/AMP: Problems Actual application of ACMG/AMP guidelines in clinical routine is limited by: Complexity Flexibility ‘‘Only 34% interpretation concordance across all sites that used the ACMGAMP guidelines’’ Amendola, Laura M. et al. “Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. ” American Journal of Human Genetics 98. 6 (2016): 1067– 1076. PMC. Web. 11 Jan. 2018. AMIA 2018 | amia. org 6
ACMG/AMP: Solution AMIA 2018 | amia. org 7
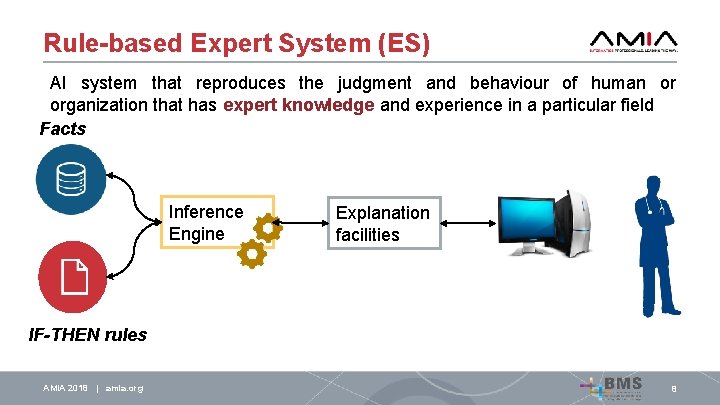
Rule-based Expert System (ES) AI system that reproduces the judgment and behaviour of human or organization that has expert knowledge and experience in a particular field Facts Inference Engine Explanation facilities IF-THEN rules AMIA 2018 | amia. org 8
Rule-based Expert System (ES) Facts IF-THEN rules AMIA 2018 | amia. org 16
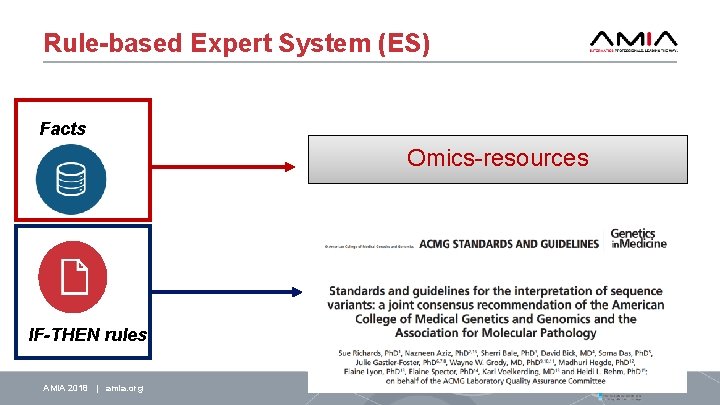
Rule-based Expert System (ES) Facts Omics-resources IF-THEN rules AMIA 2018 | amia. org 17
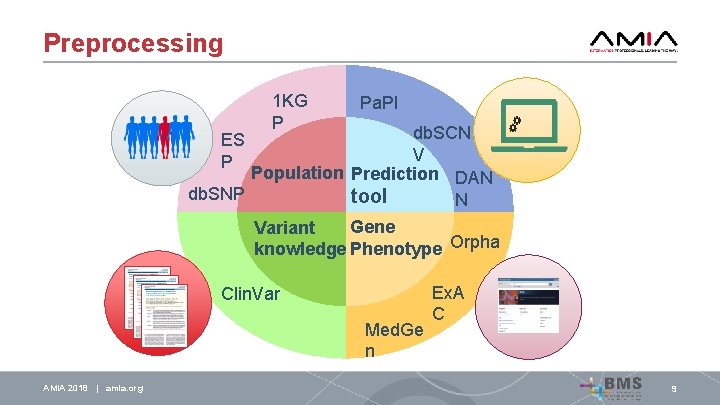
Preprocessing 1 KG P Pa. PI db. SCN V Population Prediction DAN db. SNP tool N ES P Gene Variant knowledge Phenotype Orpha Clin. Var Med. Ge n AMIA 2018 | amia. org Ex. A C 9

ACMG/AMP Criteria Implementation Pathogenic Very Strong PVS 1: «Null variant (nonsense, frameshift, etc. ) in gene where Loss of Function (LOF) is a known mechanism of disease» AMIA 2018 | amia. org 10
ACMG/AMP Criteria Implementation Pathogenic Very Strong PVS 1: «Null variant (nonsense, frameshift, etc. ) in gene where Loss of Function (LOF) is a known mechanism of disease» AMIA 2018 | amia. org ü Variant is stop gained/frameshift/canonical splicing site. ü Gene is a known LOF for a certain phenotype or Ex. AC Prec >= 0. 9, Ex. AC Pli >= 0. 9 in case of recessive and dominant phenotype respectively. ü Another pathogenic null variant in the same exon has been reported in Clin. Var. 10
ACMG/AMP Criteria Implementation Pathogenic Strong PS 3: «Well-established in vitro or in vivo functional studies supportive of damaging effect on the gene or gene product» AMIA 2018 | amia. org 11
ACMG/AMP Criteria Implementation Pathogenic Strong PS 3: «Well-established in vitro or in vivo functional studies supportive of damaging effect on the gene or gene product» AMIA 2018 | amia. org ü Variant has been reported as pathogenic for the same phenotype in Clin. Var ü the submission methods report the presence of functional studies 11

ACMG/AMP Rules IF 1 VERY STRONG (PVS 1) AND >=1 STRONG (PS 1 -PS 4) AMIA 2018 | amia. org 12
ACMG/AMP Rules IF 1 VERY STRONG (PVS 1) AND >=1 STRONG (PS 1 -PS 4) AMIA 2018 | amia. org Variant is Pathogenic 12
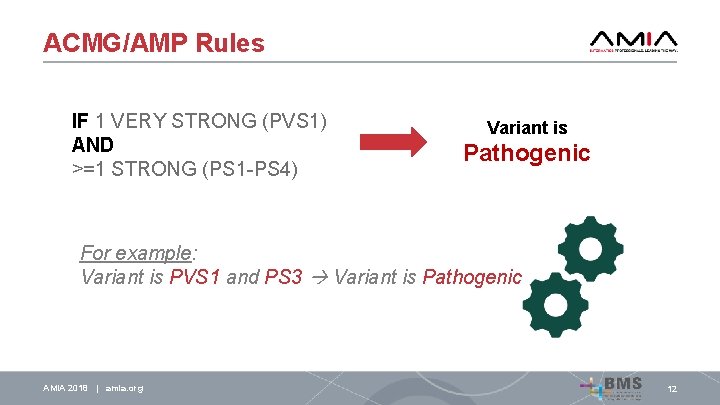
ACMG/AMP Rules IF 1 VERY STRONG (PVS 1) AND >=1 STRONG (PS 1 -PS 4) Variant is Pathogenic For example: Variant is PVS 1 and PS 3 Variant is Pathogenic AMIA 2018 | amia. org 12
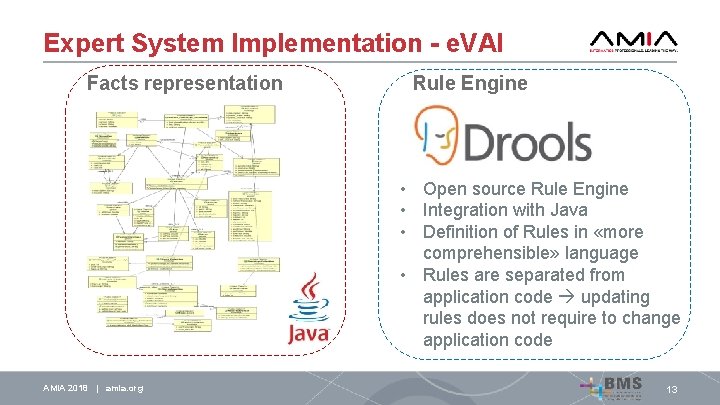
Expert System Implementation - e. VAI Facts representation Rule Engine • Open source Rule Engine • Integration with Java • Definition of Rules in «more comprehensible» language • Rules are separated from application code updating rules does not require to change application code AMIA 2018 | amia. org 13
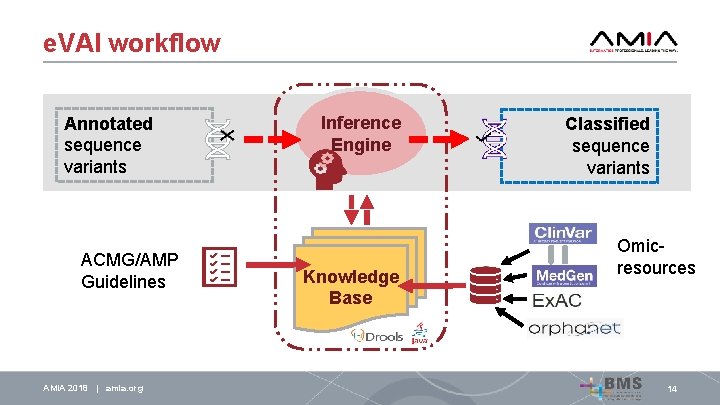
e. VAI workflow Annotated sequence variants ACMG/AMP Guidelines AMIA 2018 | amia. org Inference Engine Knowledge Base Classified sequence variants Omicresources 14
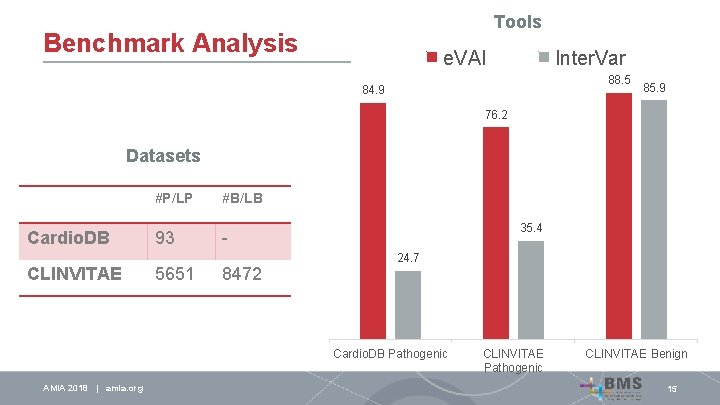
Tools Benchmark Analysis e. VAI Inter. Var 88. 5 84. 9 85. 9 76. 2 Datasets Cardio. DB CLINVITAE #P/LP #B/LB 93 - 5651 8472 35. 4 24. 7 Cardio. DB Pathogenic AMIA 2018 | amia. org CLINVITAE Pathogenic CLINVITAE Benign 15
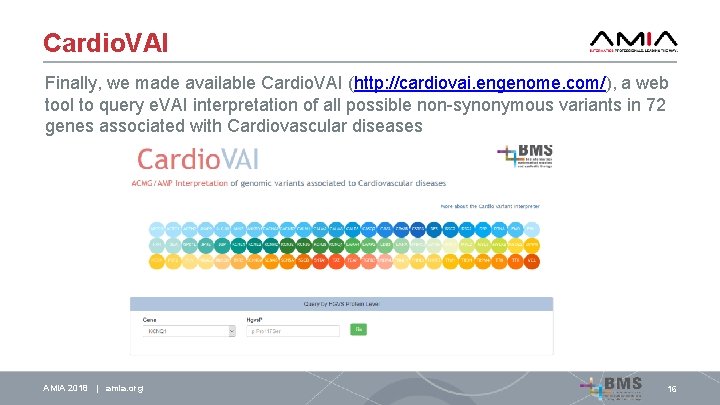
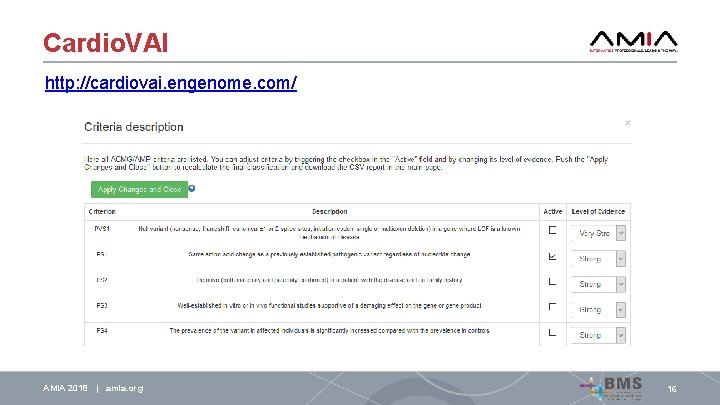
Cardio. VAI Finally, we made available Cardio. VAI (http: //cardiovai. engenome. com/), a web tool to query e. VAI interpretation of all possible non-synonymous variants in 72 genes associated with Cardiovascular diseases AMIA 2018 | amia. org 16
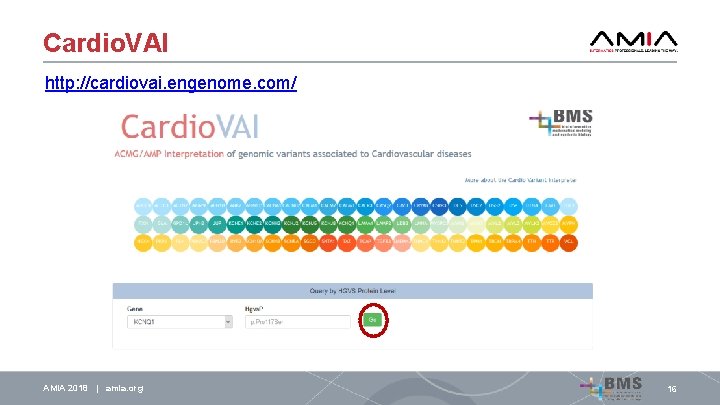
Cardio. VAI http: //cardiovai. engenome. com/ AMIA 2018 | amia. org 16

Cardio. VAI http: //cardiovai. engenome. com/ AMIA 2018 | amia. org 16
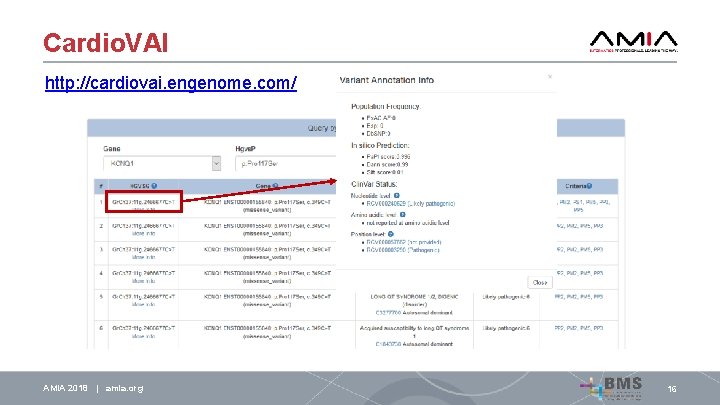
Cardio. VAI http: //cardiovai. engenome. com/ AMIA 2018 | amia. org 16
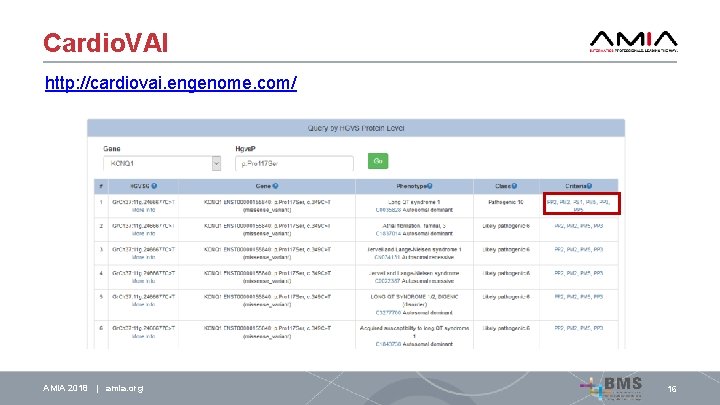
Cardio. VAI http: //cardiovai. engenome. com/ AMIA 2018 | amia. org 16
Cardio. VAI http: //cardiovai. engenome. com/ AMIA 2018 | amia. org 16

Cardio. VAI http: //cardiovai. engenome. com/ AMIA 2018 | amia. org 16

Conclusion Germline sequence variants play a role in the development of Mendelian disorders AMIA 2018 | amia. org 17

Conclusion Germline sequence variants play a role in the development of Mendelian disorders Their interpretation in clinical routine should be standardized AMIA 2018 | amia. org 17

Conclusion Germline sequence variants play a role in the development of Mendelian disorders Their interpretation in clinical routine should be standardized e. VAI is an automatic tool that supports guidelines-based variant interpretation in clinical practice AMIA 2018 | amia. org 17

Thank you! Email me at: giovanna. nicora 01@universitadipavia. it Acknowledgements Maugeri University Hospital Silvia Priori Patrick Gambelli Mirella Memmi Carlo Napolitano Ivan Limongelli, Ph. D, Prof. Riccardo Bellazzi, en. Genome srl Ph. D, University of Pavia Alberto Malovini Andrea Mazzanti
- Slides: 39