Amino Acids and the Peptide Bond Lance Wells

Amino Acids and the Peptide Bond Lance Wells Biochemistry & Molecular Biology, CCRC lwells@ccrc. uga. edu All lecture slides and papers for Thursday are available at: http: //cell. ccrc. uga. edu/~glycobiology/

Learning Objectives: • Know your 20 standard AA’s • Be able to classify based on side chain: ionizable, aliphatic, aromatic, sulfurcontaining, polar, non-polar, etc. . • Be able to draw a peptide bond and understand the amide character • Be aware of PTM’s • Appreciate that function of proteins are dictated in large part by side chain properties of AA’s
Central Dogma of Molecular Biology http: //www. labgrab. com/users/labgrab /blog/ central-dogma-geneticsincomplete_id%3 D 904
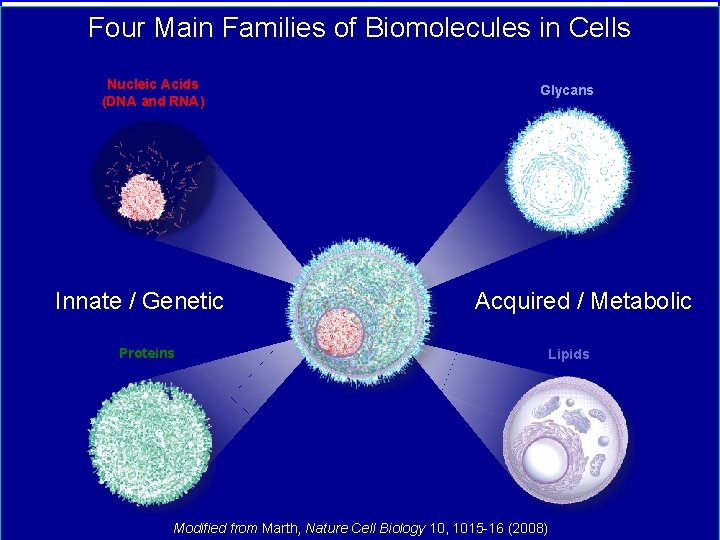
Four Main Families of Biomolecules in Cells Nucleic Acids (DNA and RNA) Innate / Genetic Glycans Acquired / Metabolic Proteins Modified from Marth, Nature Cell Biology 10, 1015 -16 (2008) Lipids
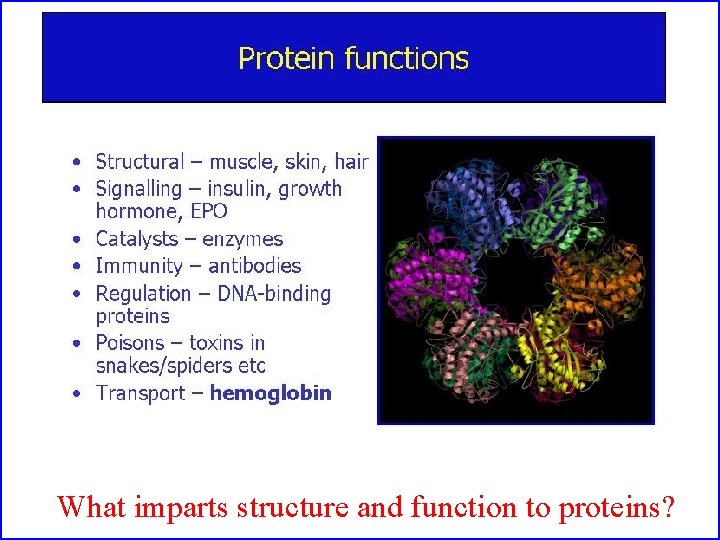
What imparts structure and function to proteins?
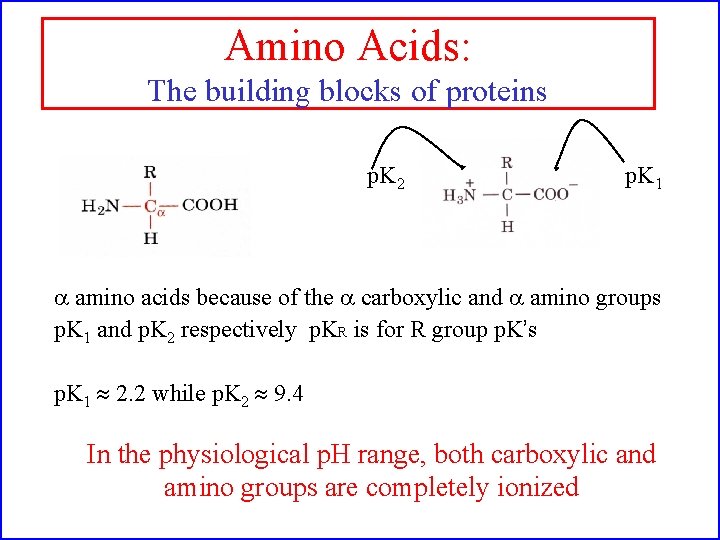
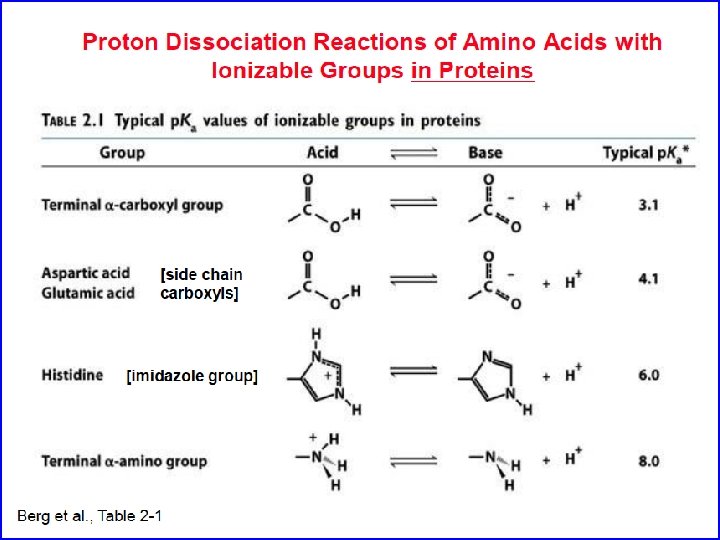
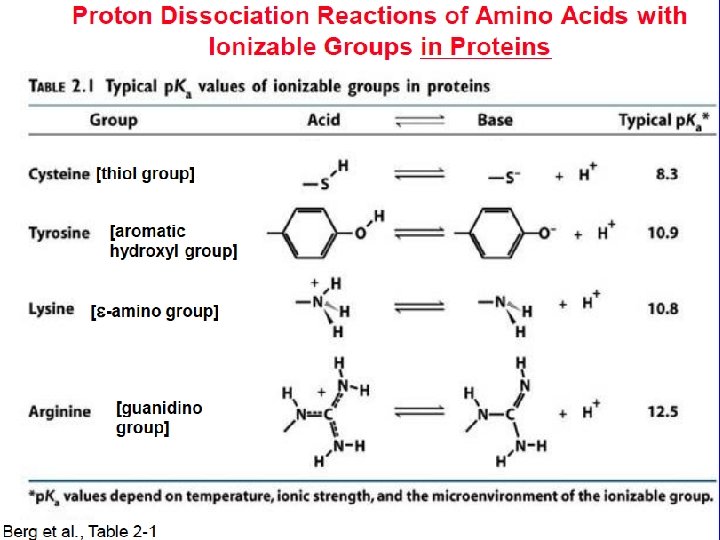
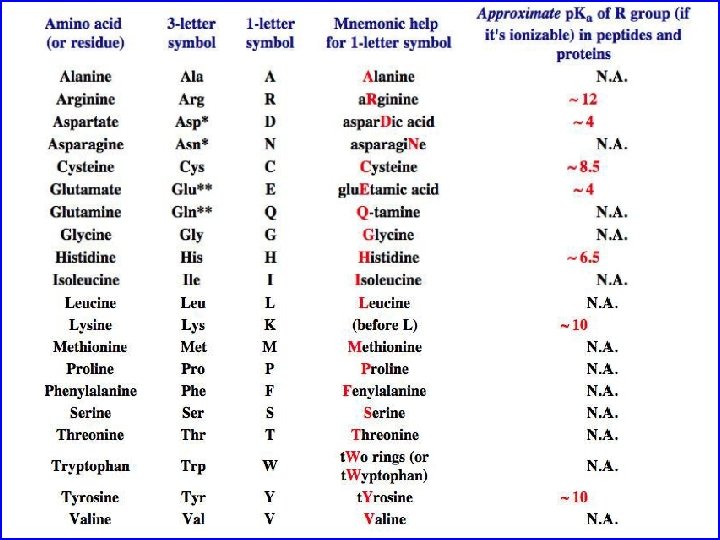
Amino Acids: The building blocks of proteins p. K 2 p. K 1 a amino acids because of the a carboxylic and a amino groups p. K 1 and p. K 2 respectively p. KR is for R group p. K’s p. K 1 2. 2 while p. K 2 9. 4 In the physiological p. H range, both carboxylic and amino groups are completely ionized

Amino acids are Ampholytes They can act as either an acid or a base They are Zwitterions or molecules that have both a positive and a negative charge
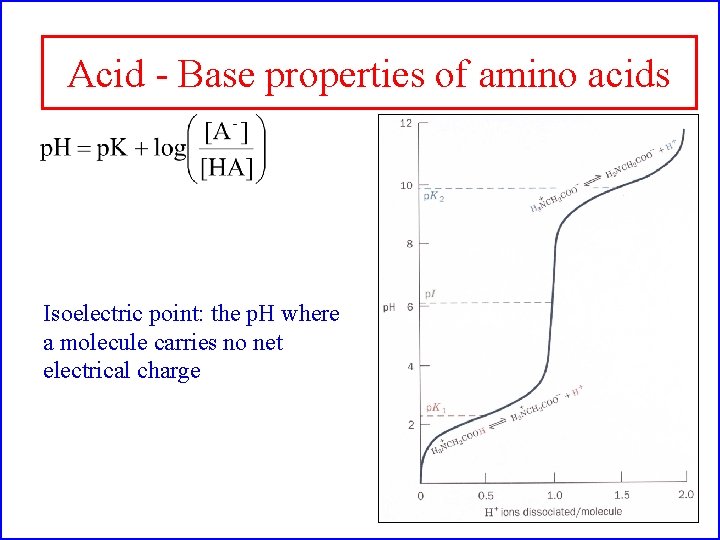
Acid - Base properties of amino acids Isoelectric point: the p. H where a molecule carries no net electrical charge
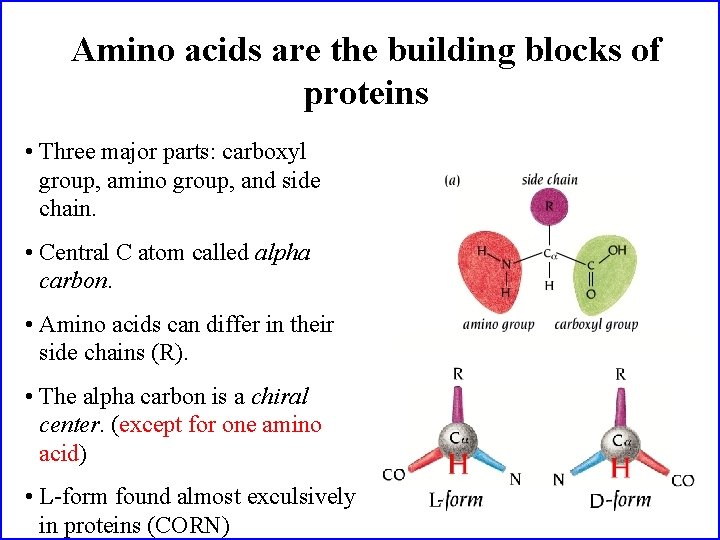
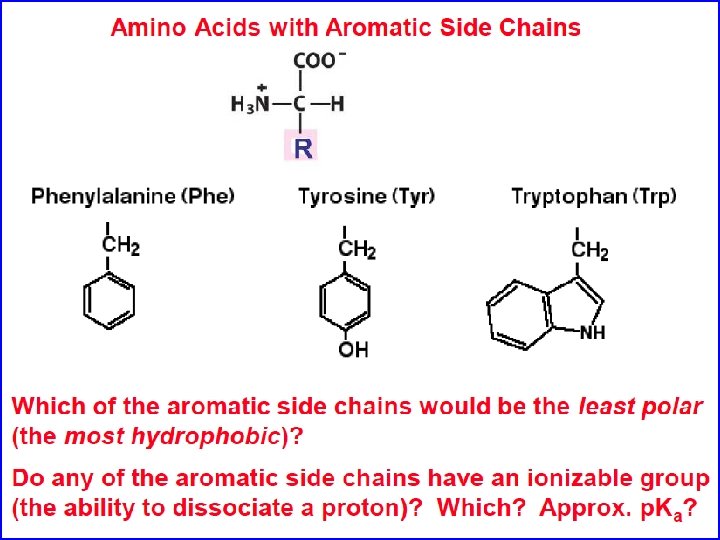
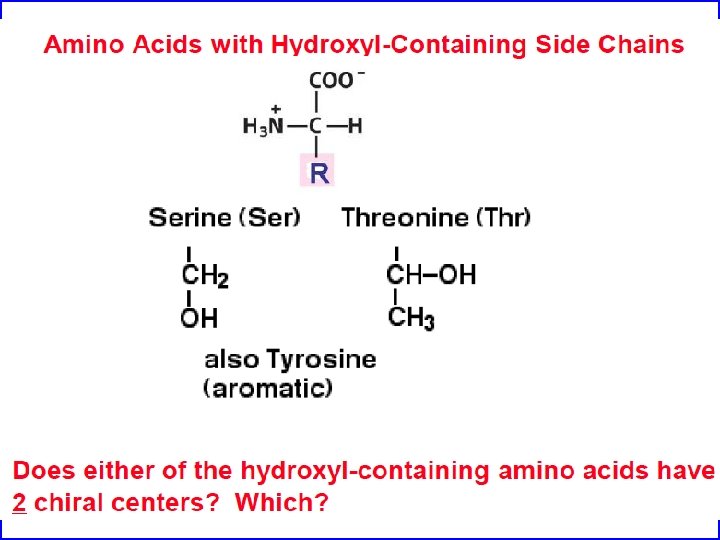
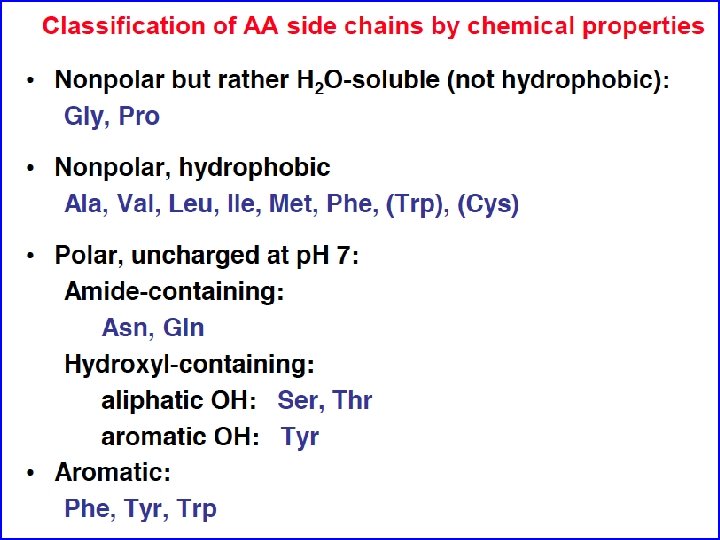
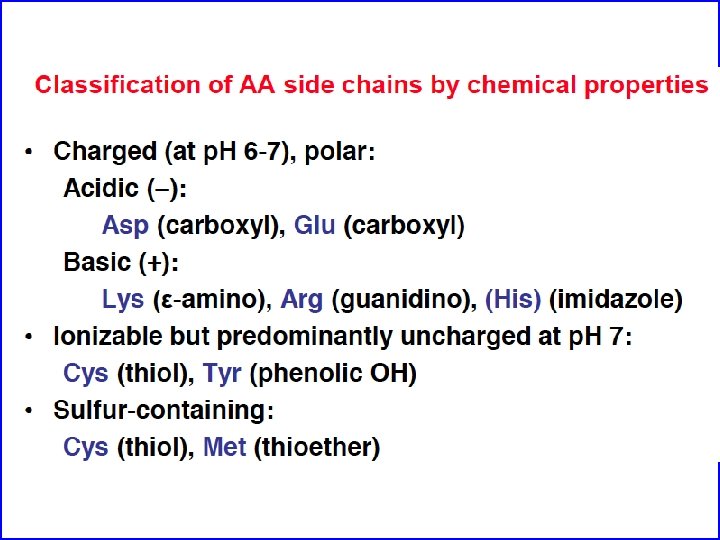
Amino acids are the building blocks of proteins • Three major parts: carboxyl group, amino group, and side chain. • Central C atom called alpha carbon. • Amino acids can differ in their side chains (R). • The alpha carbon is a chiral center. (except for one amino acid) • L-form found almost exculsively in proteins (CORN)
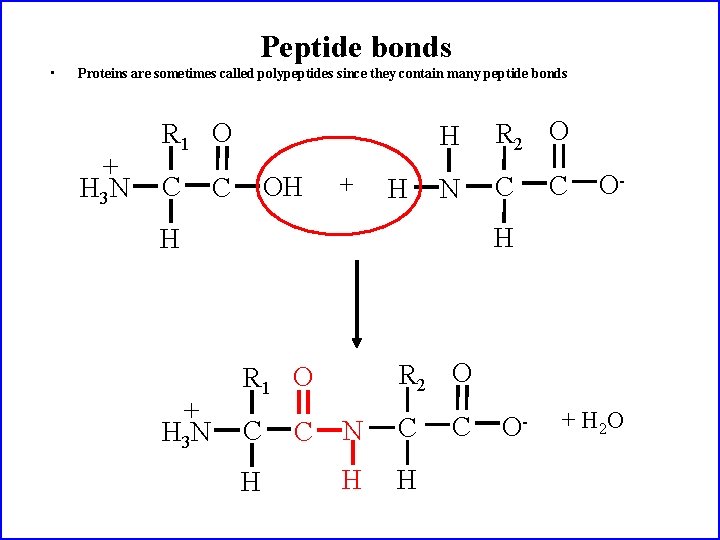
Peptide bonds • Proteins are sometimes called polypeptides since they contain many peptide bonds + H 3 N R 1 O C OH C + H H R 2 O N C O- H H + H 3 N C R 2 O R 1 O C H C N C H H C O- + H 2 O
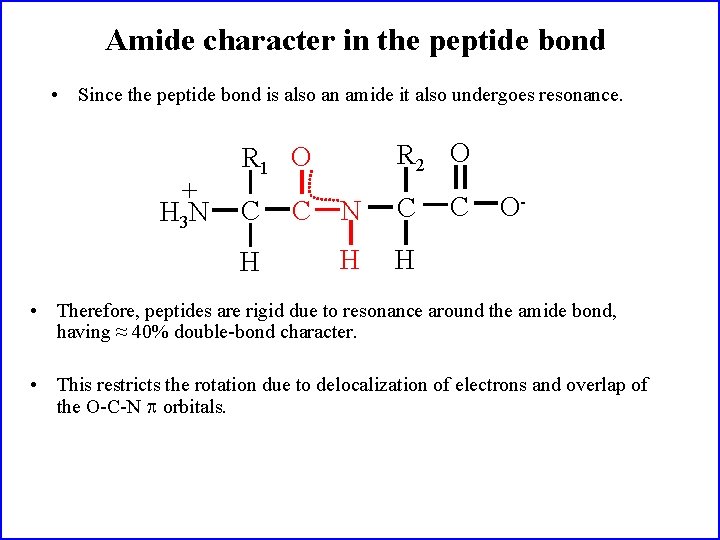
Amide character in the peptide bond • Since the peptide bond is also an amide it also undergoes resonance. + H 3 N R 2 O R 1 O C H C N C H H C O- • Therefore, peptides are rigid due to resonance around the amide bond, having ≈ 40% double-bond character. • This restricts the rotation due to delocalization of electrons and overlap of the O-C-N orbitals.
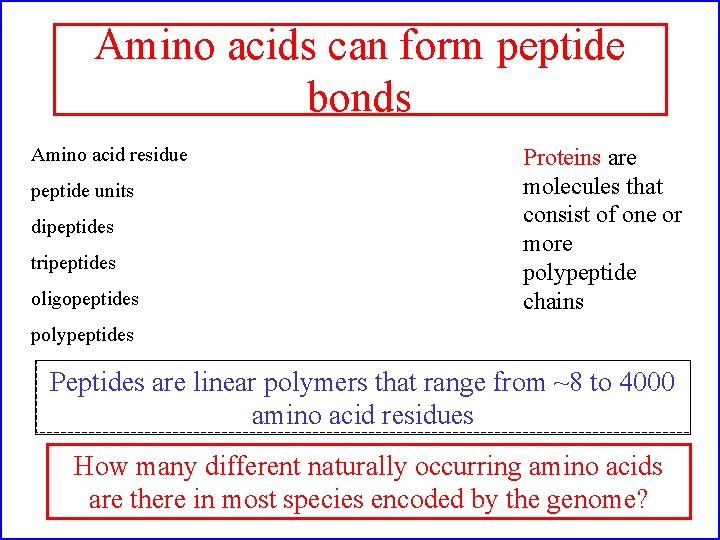
Amino acids can form peptide bonds Amino acid residue peptide units dipeptides tripeptides oligopeptides Proteins are molecules that consist of one or more polypeptide chains polypeptides Peptides are linear polymers that range from ~8 to 4000 amino acid residues How many different naturally occurring amino acids are there in most species encoded by the genome?
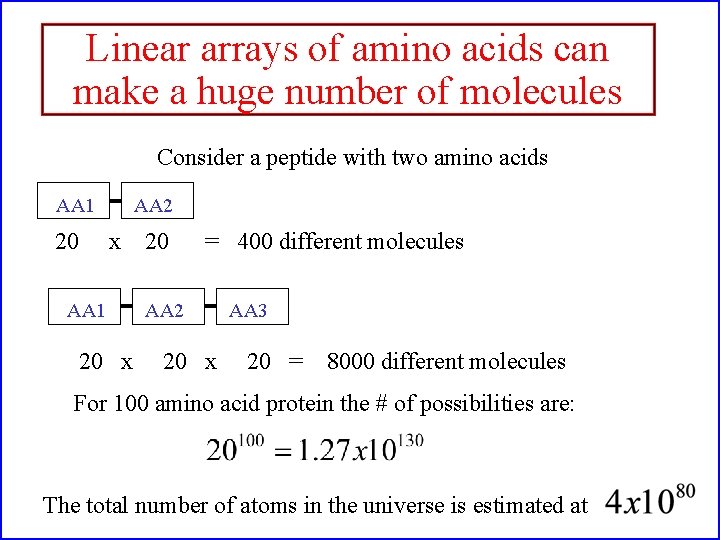
Linear arrays of amino acids can make a huge number of molecules Consider a peptide with two amino acids AA 1 20 AA 2 x AA 1 20 x 20 = 400 different molecules AA 2 20 x AA 3 20 = 8000 different molecules For 100 amino acid protein the # of possibilities are: The total number of atoms in the universe is estimated at
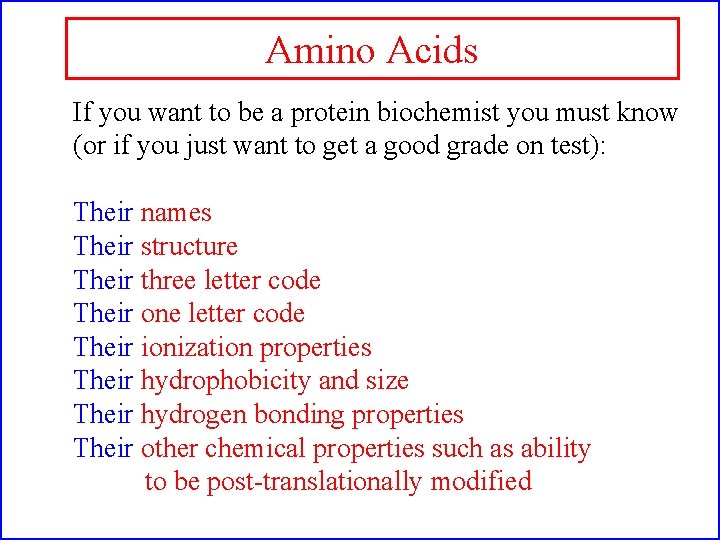
Amino Acids If you want to be a protein biochemist you must know (or if you just want to get a good grade on test): Their names Their structure Their three letter code Their one letter code Their ionization properties Their hydrophobicity and size Their hydrogen bonding properties Their other chemical properties such as ability to be post-translationally modified

Why you need to know your AA’s and their properties?

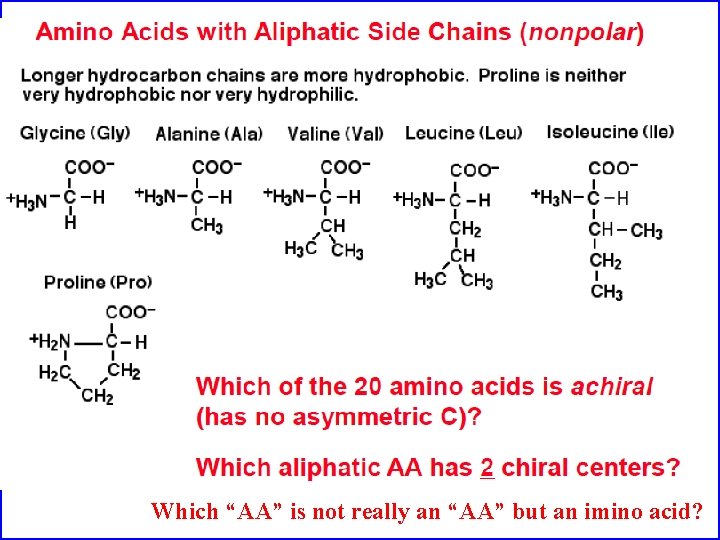
Which “AA” is not really an “AA” but an imino acid?
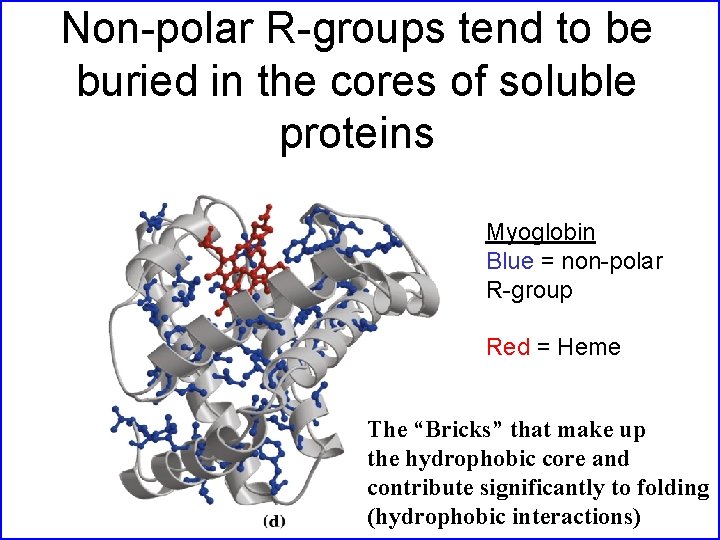
Non-polar R-groups tend to be buried in the cores of soluble proteins Myoglobin Blue = non-polar R-group Red = Heme The “Bricks” that make up the hydrophobic core and contribute significantly to folding (hydrophobic interactions)
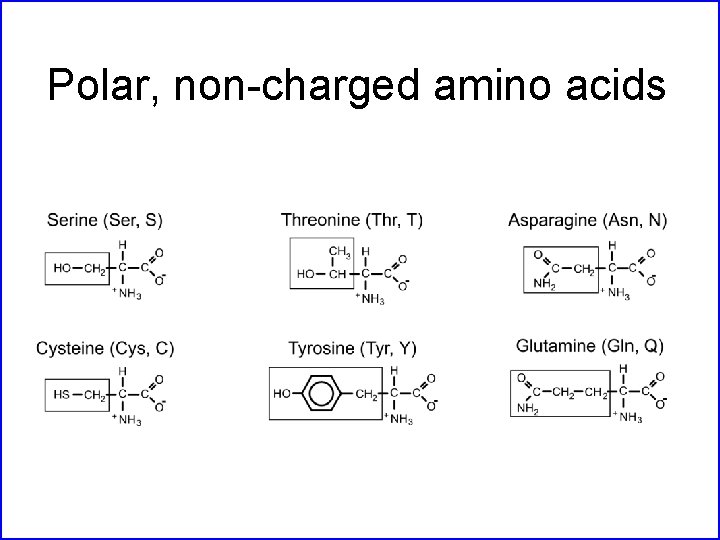
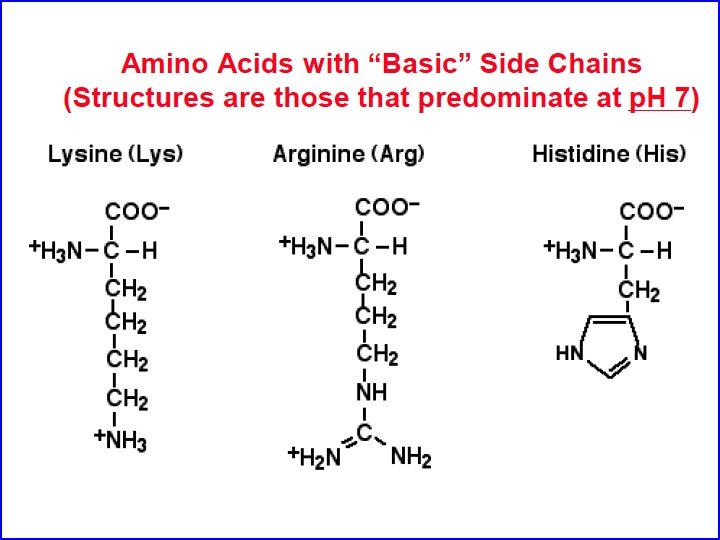
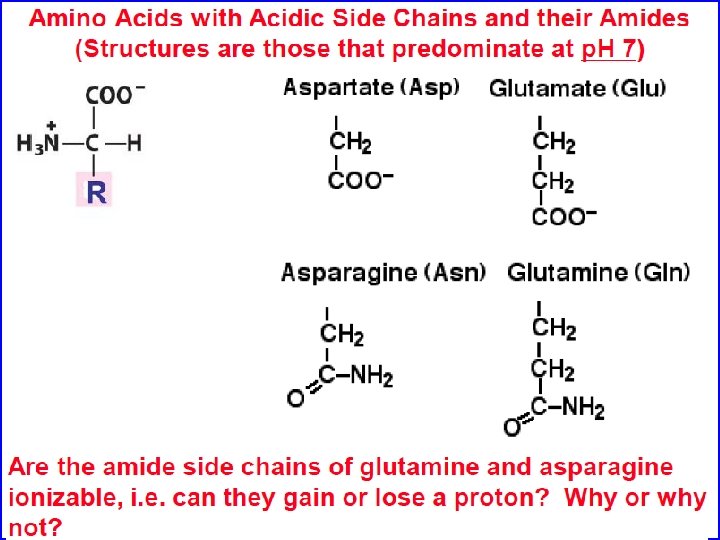
Polar, non-charged amino acids
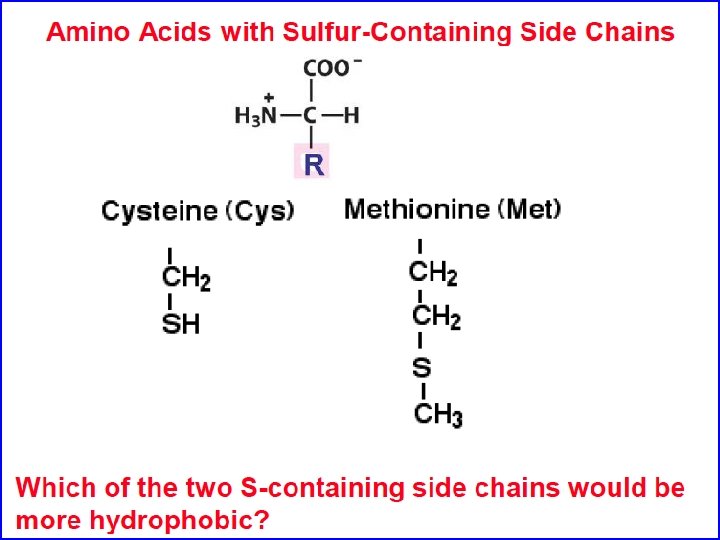
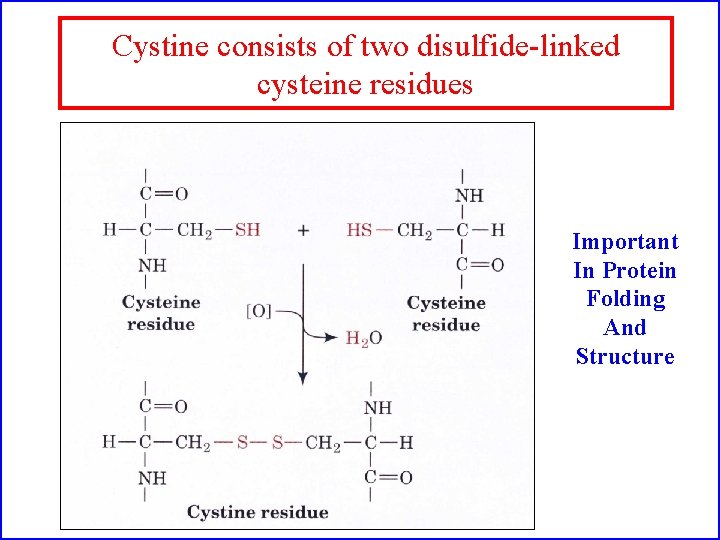
Cystine consists of two disulfide-linked cysteine residues Important In Protein Folding And Structure
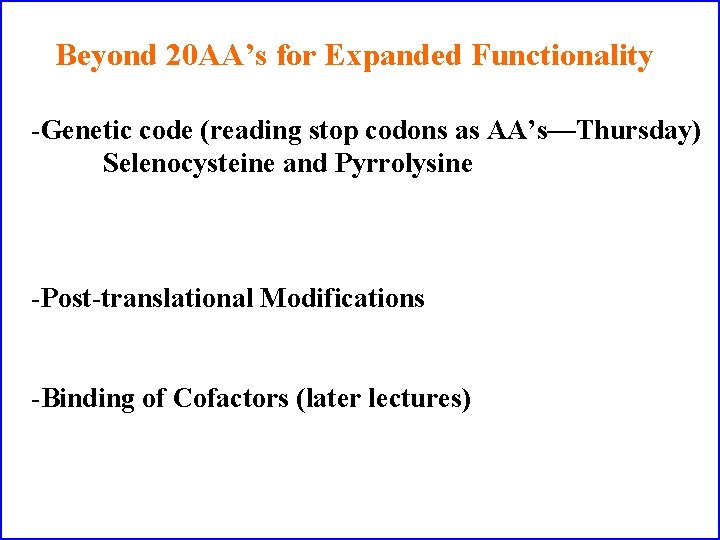
Beyond 20 AA’s for Expanded Functionality -Genetic code (reading stop codons as AA’s—Thursday) Selenocysteine and Pyrrolysine -Post-translational Modifications -Binding of Cofactors (later lectures)
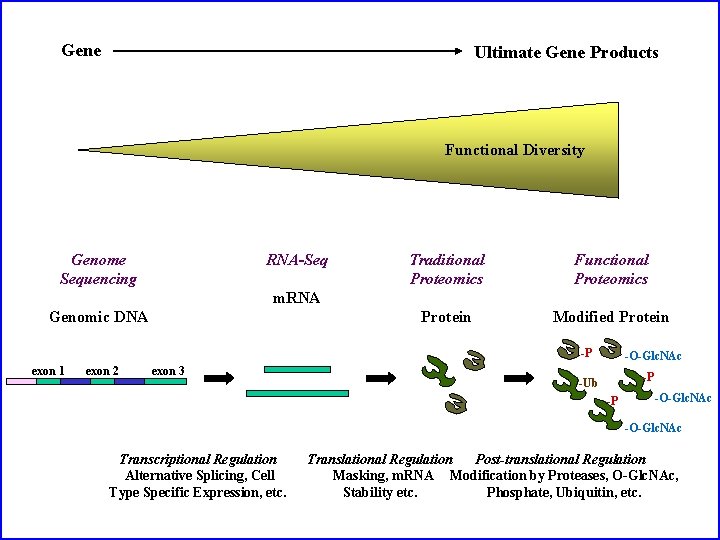
Gene Ultimate Gene Products Functional Diversity Genome Sequencing RNA-Seq Traditional Proteomics Functional Proteomics Protein Modified Protein m. RNA Genomic DNA -P exon 1 exon 2 exon 3 -O-Glc. NAc -P -Ub -P -O-Glc. NAc Transcriptional Regulation Alternative Splicing, Cell Type Specific Expression, etc. Translational Regulation Post-translational Regulation Masking, m. RNA Modification by Proteases, O-Glc. NAc, Stability etc. Phosphate, Ubiquitin, etc.

Lots, and lots, of PTMs

Post-translational Modification of Amino Acids (just a few examples of impact) Phosphorylation (Typically Ser, Thr, Tyr) Glycosylation (Typically Asn, Ser, Thr) gain of charge, binding solubility, stability, binding Acetylation/Acylation/Methylation (N-term, Lys, Arg) Lipidation/prenylation (Typically Cys) Ubiquitination/Sumoylation (Lys) loss of charge, stability membrane anchoring degradation, sign
- Slides: 34