Amino acid activation The twostep process in which

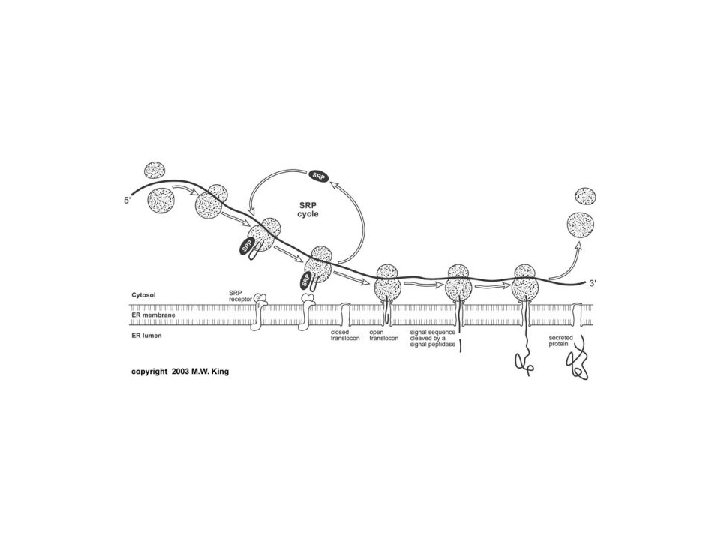


- Slides: 33

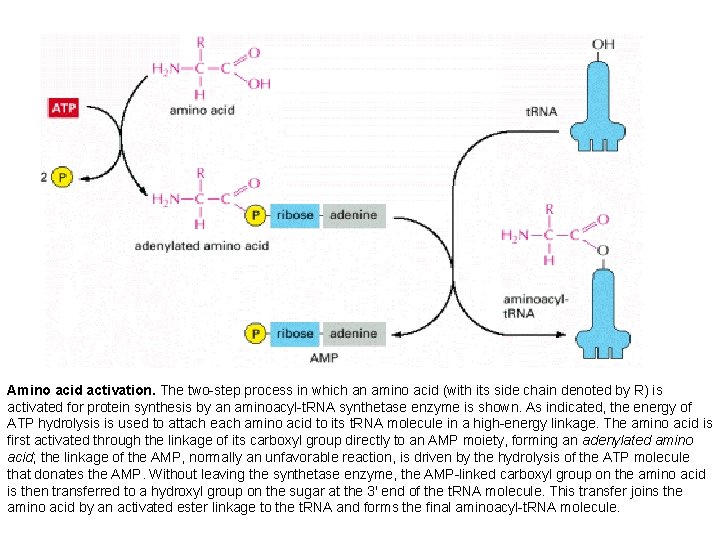
Amino acid activation. The two-step process in which an amino acid (with its side chain denoted by R) is activated for protein synthesis by an aminoacyl-t. RNA synthetase enzyme is shown. As indicated, the energy of ATP hydrolysis is used to attach each amino acid to its t. RNA molecule in a high-energy linkage. The amino acid is first activated through the linkage of its carboxyl group directly to an AMP moiety, forming an adenylated amino acid; the linkage of the AMP, normally an unfavorable reaction, is driven by the hydrolysis of the ATP molecule that donates the AMP. Without leaving the synthetase enzyme, the AMP-linked carboxyl group on the amino acid is then transferred to a hydroxyl group on the sugar at the 3′ end of the t. RNA molecule. This transfer joins the amino acid by an activated ester linkage to the t. RNA and forms the final aminoacyl-t. RNA molecule.

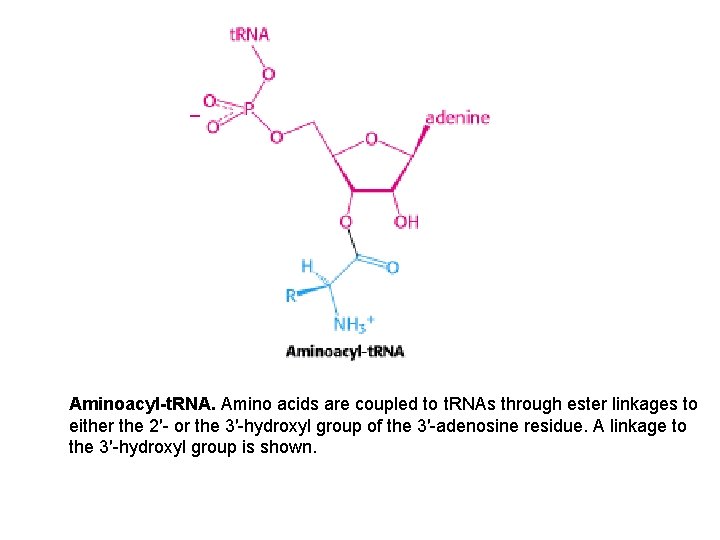
Aminoacyl-t. RNA. Amino acids are coupled to t. RNAs through ester linkages to either the 2′- or the 3′-hydroxyl group of the 3′-adenosine residue. A linkage to the 3′-hydroxyl group is shown.

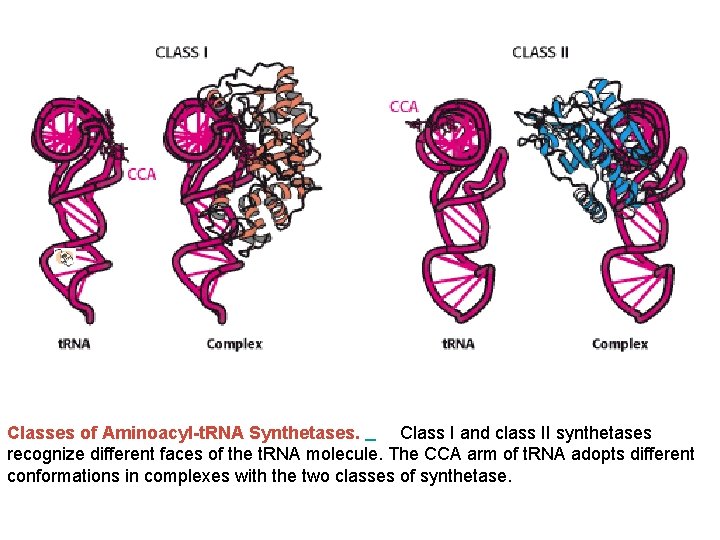
Classes of Aminoacyl-t. RNA Synthetases. Class I and class II synthetases recognize different faces of the t. RNA molecule. The CCA arm of t. RNA adopts different conformations in complexes with the two classes of synthetase.

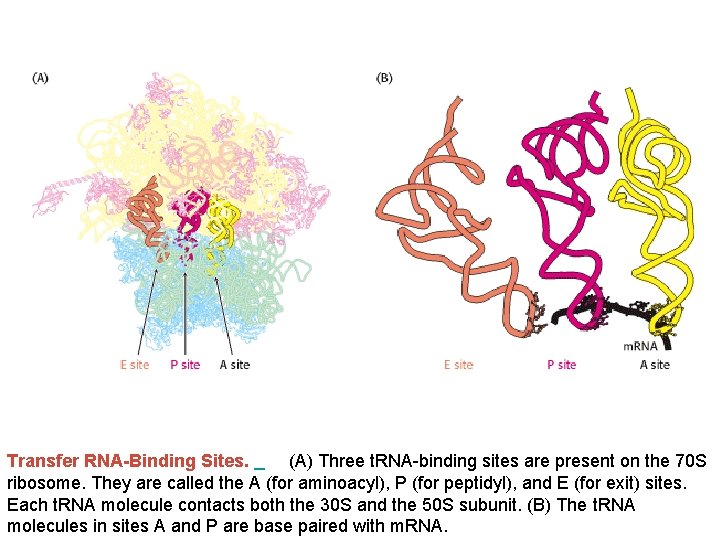
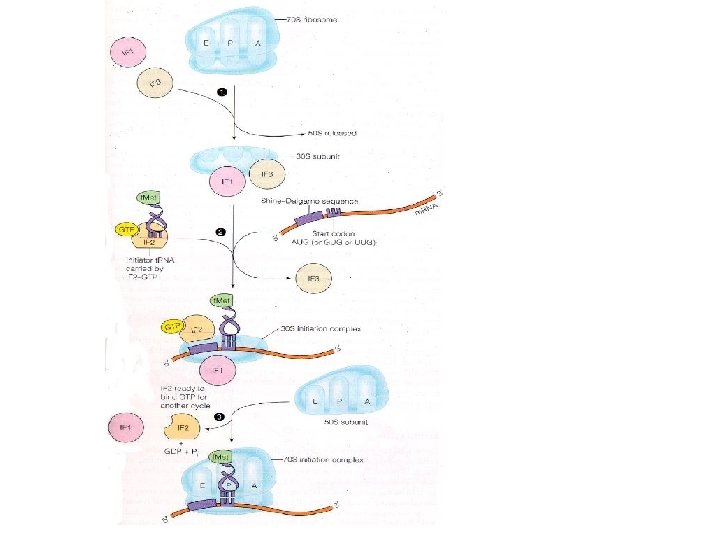
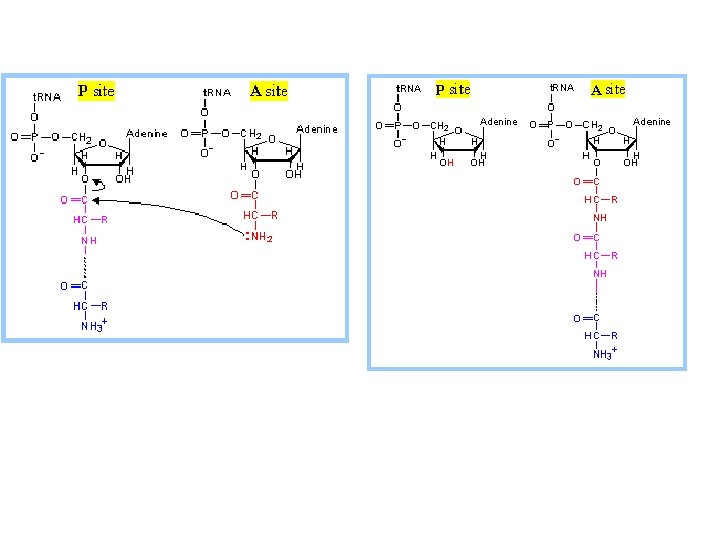
Transfer RNA-Binding Sites. (A) Three t. RNA-binding sites are present on the 70 S ribosome. They are called the A (for aminoacyl), P (for peptidyl), and E (for exit) sites. Each t. RNA molecule contacts both the 30 S and the 50 S subunit. (B) The t. RNA molecules in sites A and P are base paired with m. RNA.

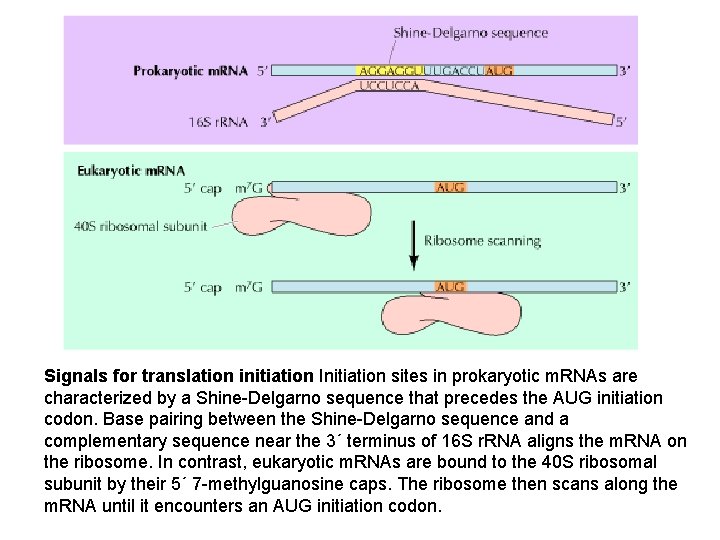
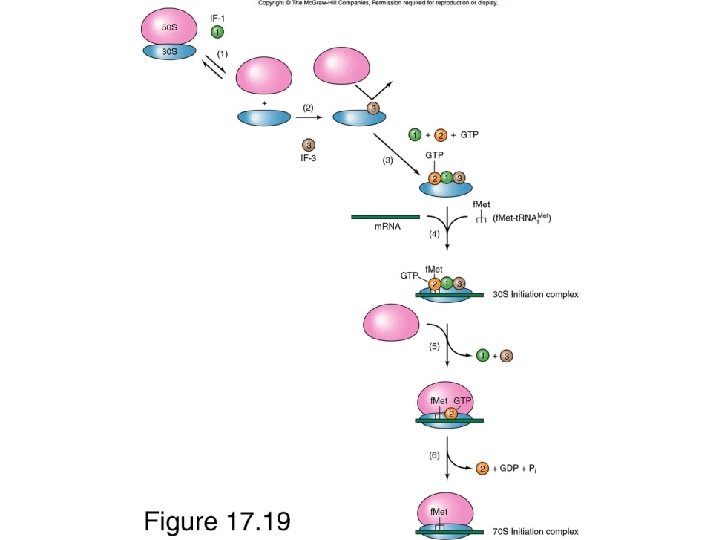
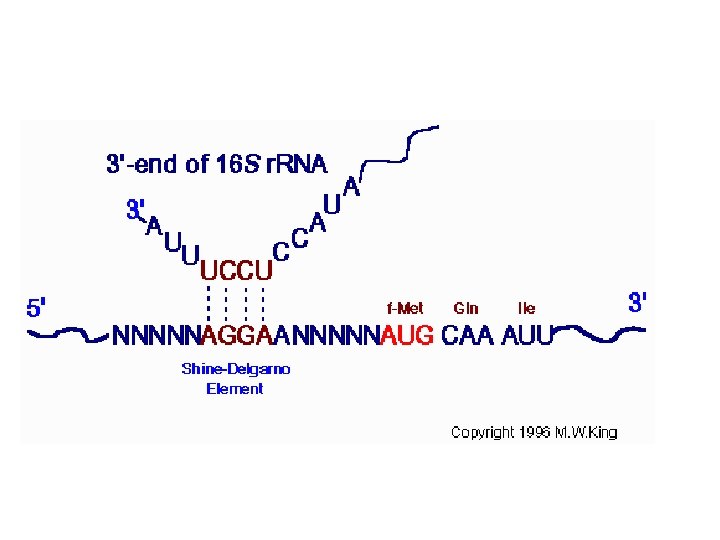
Signals for translation initiation Initiation sites in prokaryotic m. RNAs are characterized by a Shine-Delgarno sequence that precedes the AUG initiation codon. Base pairing between the Shine-Delgarno sequence and a complementary sequence near the 3´ terminus of 16 S r. RNA aligns the m. RNA on the ribosome. In contrast, eukaryotic m. RNAs are bound to the 40 S ribosomal subunit by their 5´ 7 -methylguanosine caps. The ribosome then scans along the m. RNA until it encounters an AUG initiation codon.


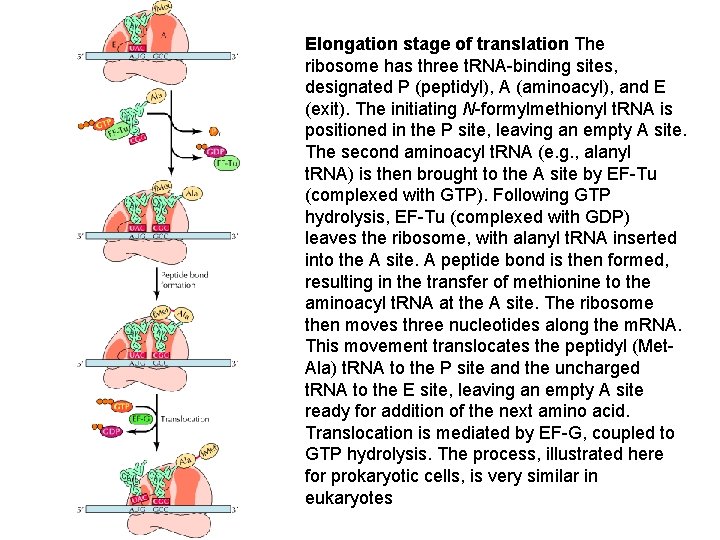
Elongation stage of translation The ribosome has three t. RNA-binding sites, designated P (peptidyl), A (aminoacyl), and E (exit). The initiating N-formylmethionyl t. RNA is positioned in the P site, leaving an empty A site. The second aminoacyl t. RNA (e. g. , alanyl t. RNA) is then brought to the A site by EF-Tu (complexed with GTP). Following GTP hydrolysis, EF-Tu (complexed with GDP) leaves the ribosome, with alanyl t. RNA inserted into the A site. A peptide bond is then formed, resulting in the transfer of methionine to the aminoacyl t. RNA at the A site. The ribosome then moves three nucleotides along the m. RNA. This movement translocates the peptidyl (Met. Ala) t. RNA to the P site and the uncharged t. RNA to the E site, leaving an empty A site ready for addition of the next amino acid. Translocation is mediated by EF-G, coupled to GTP hydrolysis. The process, illustrated here for prokaryotic cells, is very similar in eukaryotes


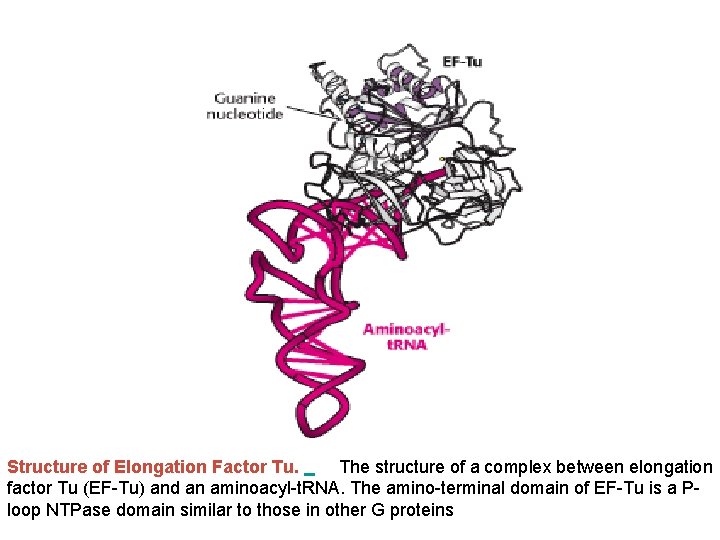
Structure of Elongation Factor Tu. The structure of a complex between elongation factor Tu (EF-Tu) and an aminoacyl-t. RNA. The amino-terminal domain of EF-Tu is a Ploop NTPase domain similar to those in other G proteins

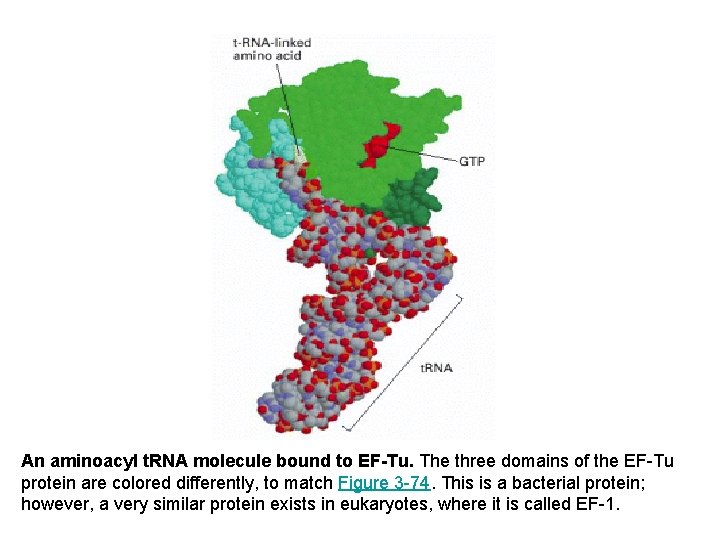
An aminoacyl t. RNA molecule bound to EF-Tu. The three domains of the EF-Tu protein are colored differently, to match Figure 3 -74. This is a bacterial protein; however, a very similar protein exists in eukaryotes, where it is called EF-1.

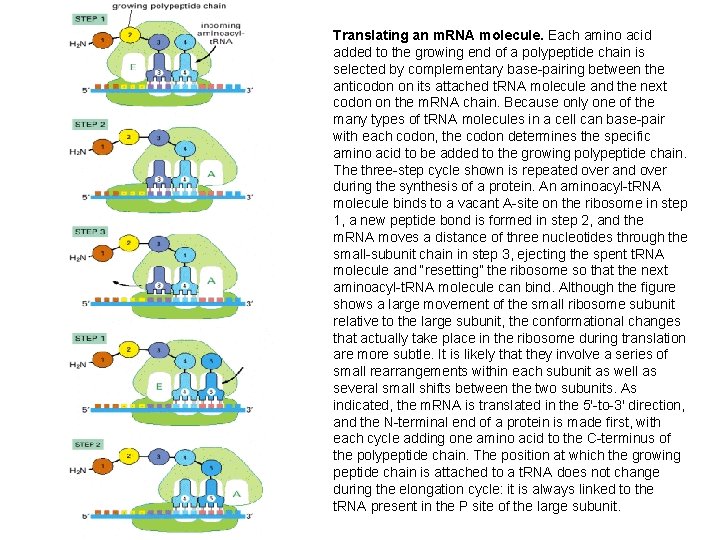
Translating an m. RNA molecule. Each amino acid added to the growing end of a polypeptide chain is selected by complementary base-pairing between the anticodon on its attached t. RNA molecule and the next codon on the m. RNA chain. Because only one of the many types of t. RNA molecules in a cell can base-pair with each codon, the codon determines the specific amino acid to be added to the growing polypeptide chain. The three-step cycle shown is repeated over and over during the synthesis of a protein. An aminoacyl-t. RNA molecule binds to a vacant A-site on the ribosome in step 1, a new peptide bond is formed in step 2, and the m. RNA moves a distance of three nucleotides through the small-subunit chain in step 3, ejecting the spent t. RNA molecule and “resetting” the ribosome so that the next aminoacyl-t. RNA molecule can bind. Although the figure shows a large movement of the small ribosome subunit relative to the large subunit, the conformational changes that actually take place in the ribosome during translation are more subtle. It is likely that they involve a series of small rearrangements within each subunit as well as several small shifts between the two subunits. As indicated, the m. RNA is translated in the 5′-to-3′ direction, and the N-terminal end of a protein is made first, with each cycle adding one amino acid to the C-terminus of the polypeptide chain. The position at which the growing peptide chain is attached to a t. RNA does not change during the elongation cycle: it is always linked to the t. RNA present in the P site of the large subunit.


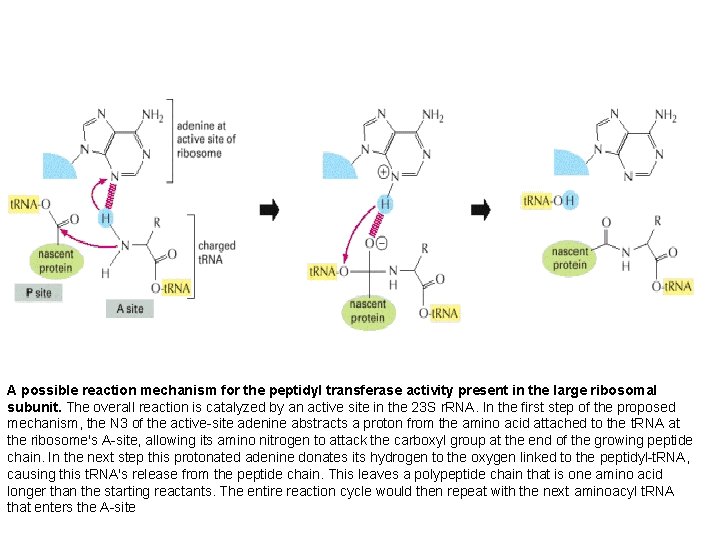
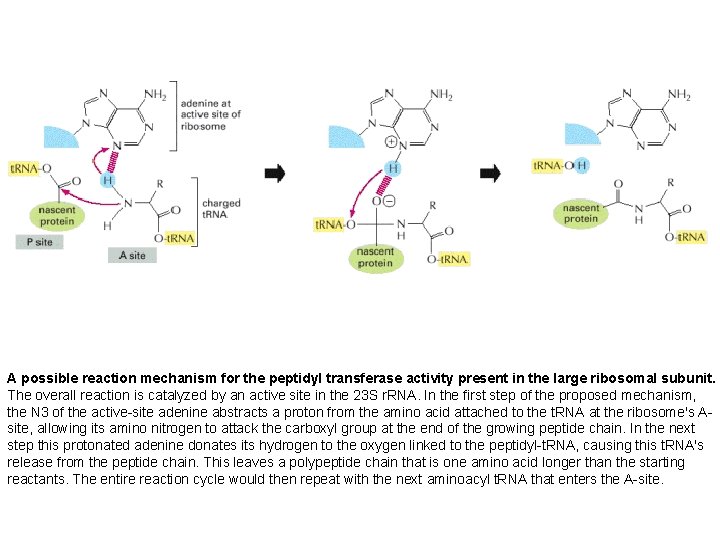
A possible reaction mechanism for the peptidyl transferase activity present in the large ribosomal subunit. The overall reaction is catalyzed by an active site in the 23 S r. RNA. In the first step of the proposed mechanism, the N 3 of the active-site adenine abstracts a proton from the amino acid attached to the t. RNA at the ribosome's A-site, allowing its amino nitrogen to attack the carboxyl group at the end of the growing peptide chain. In the next step this protonated adenine donates its hydrogen to the oxygen linked to the peptidyl-t. RNA, causing this t. RNA's release from the peptide chain. This leaves a polypeptide chain that is one amino acid longer than the starting reactants. The entire reaction cycle would then repeat with the next aminoacyl t. RNA that enters the A-site

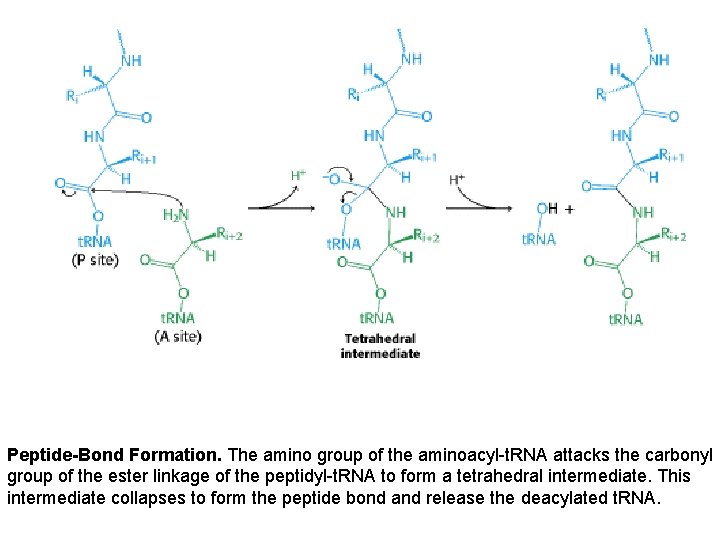
Peptide-Bond Formation. The amino group of the aminoacyl-t. RNA attacks the carbonyl group of the ester linkage of the peptidyl-t. RNA to form a tetrahedral intermediate. This intermediate collapses to form the peptide bond and release the deacylated t. RNA.

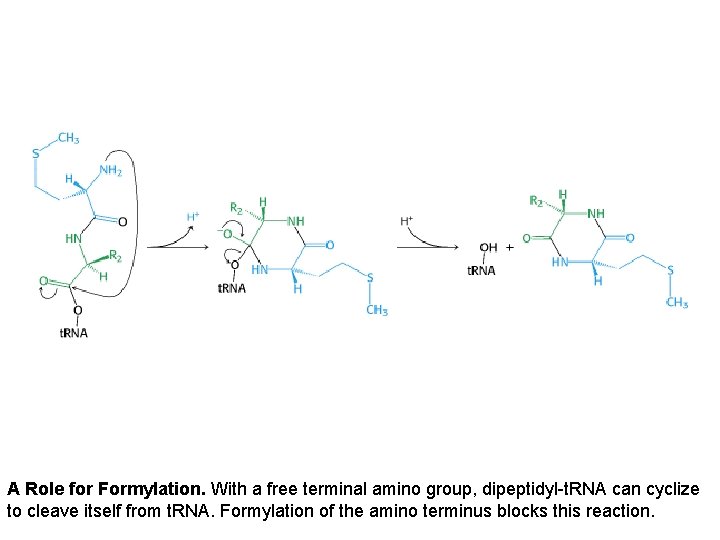
A Role for Formylation. With a free terminal amino group, dipeptidyl-t. RNA can cyclize to cleave itself from t. RNA. Formylation of the amino terminus blocks this reaction.

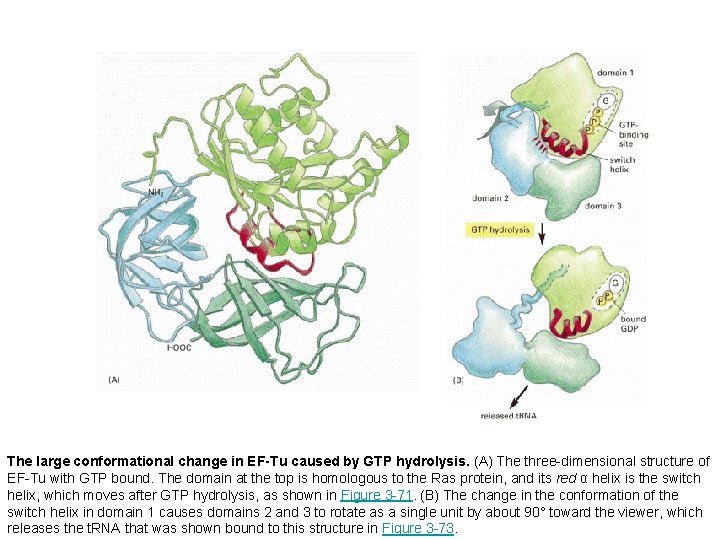
The large conformational change in EF-Tu caused by GTP hydrolysis. (A) The three-dimensional structure of EF-Tu with GTP bound. The domain at the top is homologous to the Ras protein, and its red α helix is the switch helix, which moves after GTP hydrolysis, as shown in Figure 3 -71. (B) The change in the conformation of the switch helix in domain 1 causes domains 2 and 3 to rotate as a single unit by about 90° toward the viewer, which releases the t. RNA that was shown bound to this structure in Figure 3 -73.

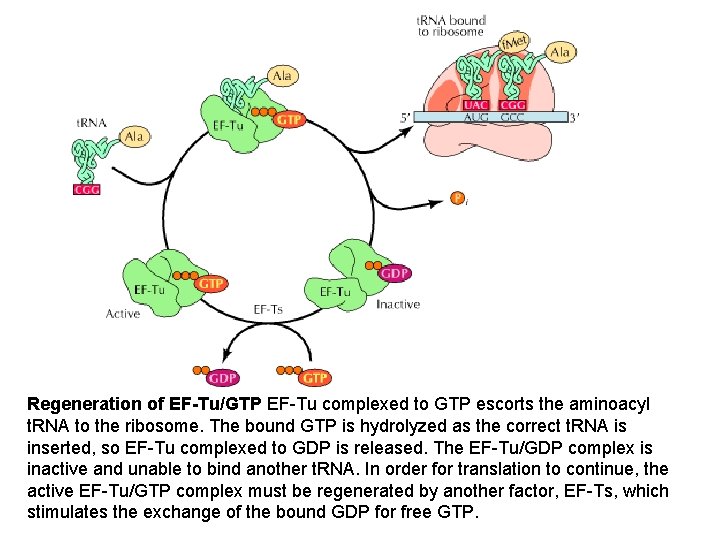
Regeneration of EF-Tu/GTP EF-Tu complexed to GTP escorts the aminoacyl t. RNA to the ribosome. The bound GTP is hydrolyzed as the correct t. RNA is inserted, so EF-Tu complexed to GDP is released. The EF-Tu/GDP complex is inactive and unable to bind another t. RNA. In order for translation to continue, the active EF-Tu/GTP complex must be regenerated by another factor, EF-Ts, which stimulates the exchange of the bound GDP for free GTP.

The final phase of protein synthesis. The binding of a release factor to an A-site bearing a stop codon terminates translation. The completed polypeptide is released and, after the action of a ribosome recycling factor (not shown), the ribosome dissociates into its two separate subunits.

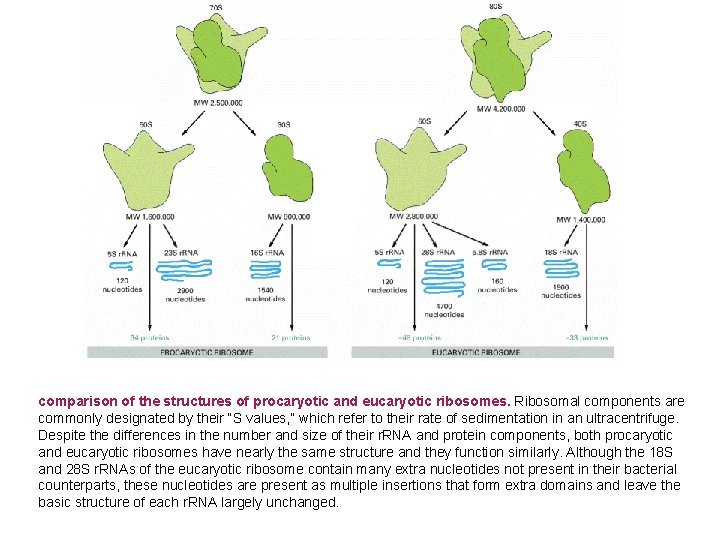
comparison of the structures of procaryotic and eucaryotic ribosomes. Ribosomal components are commonly designated by their “S values, ” which refer to their rate of sedimentation in an ultracentrifuge. Despite the differences in the number and size of their r. RNA and protein components, both procaryotic and eucaryotic ribosomes have nearly the same structure and they function similarly. Although the 18 S and 28 S r. RNAs of the eucaryotic ribosome contain many extra nucleotides not present in their bacterial counterparts, these nucleotides are present as multiple insertions that form extra domains and leave the basic structure of each r. RNA largely unchanged.

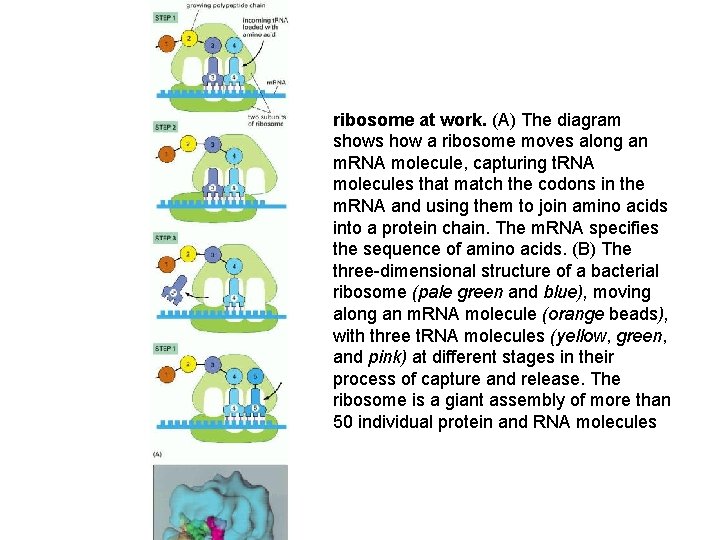
ribosome at work. (A) The diagram shows how a ribosome moves along an m. RNA molecule, capturing t. RNA molecules that match the codons in the m. RNA and using them to join amino acids into a protein chain. The m. RNA specifies the sequence of amino acids. (B) The three-dimensional structure of a bacterial ribosome (pale green and blue), moving along an m. RNA molecule (orange beads), with three t. RNA molecules (yellow, green, and pink) at different stages in their process of capture and release. The ribosome is a giant assembly of more than 50 individual protein and RNA molecules

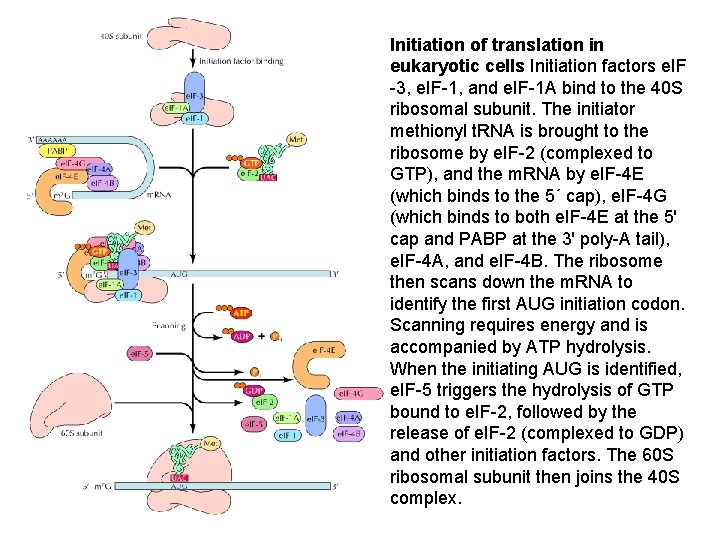
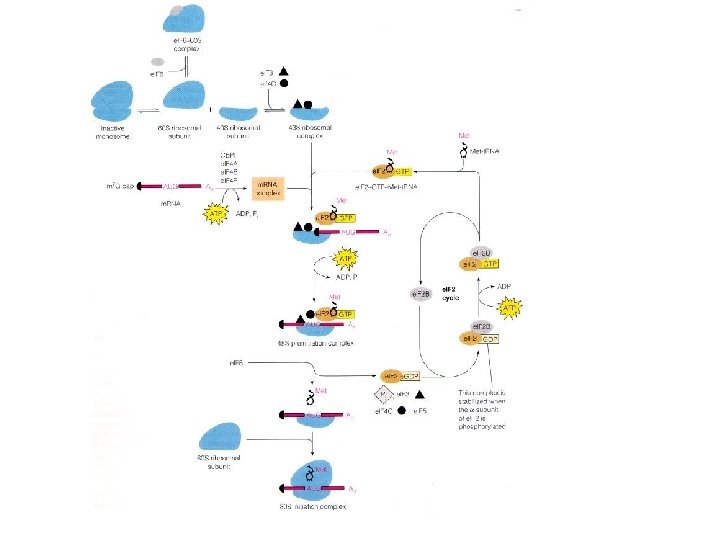
Initiation of translation in eukaryotic cells Initiation factors e. IF -3, e. IF-1, and e. IF-1 A bind to the 40 S ribosomal subunit. The initiator methionyl t. RNA is brought to the ribosome by e. IF-2 (complexed to GTP), and the m. RNA by e. IF-4 E (which binds to the 5´ cap), e. IF-4 G (which binds to both e. IF-4 E at the 5' cap and PABP at the 3' poly-A tail), e. IF-4 A, and e. IF-4 B. The ribosome then scans down the m. RNA to identify the first AUG initiation codon. Scanning requires energy and is accompanied by ATP hydrolysis. When the initiating AUG is identified, e. IF-5 triggers the hydrolysis of GTP bound to e. IF-2, followed by the release of e. IF-2 (complexed to GDP) and other initiation factors. The 60 S ribosomal subunit then joins the 40 S complex.

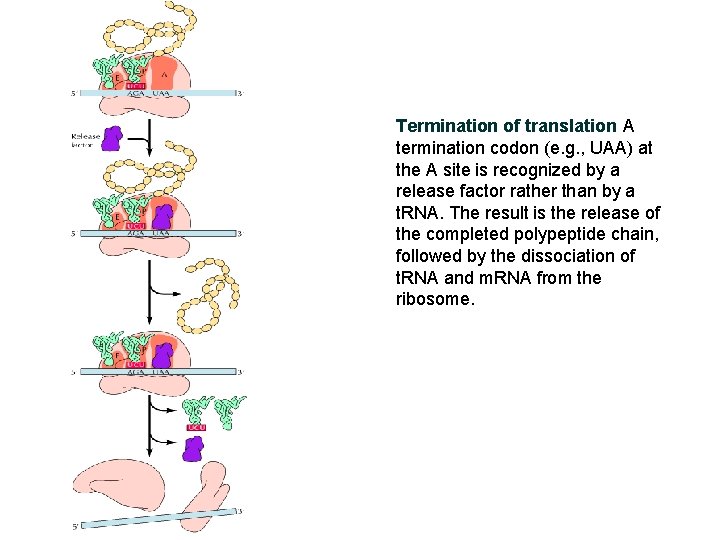
Termination of translation A termination codon (e. g. , UAA) at the A site is recognized by a release factor rather than by a t. RNA. The result is the release of the completed polypeptide chain, followed by the dissociation of t. RNA and m. RNA from the ribosome.



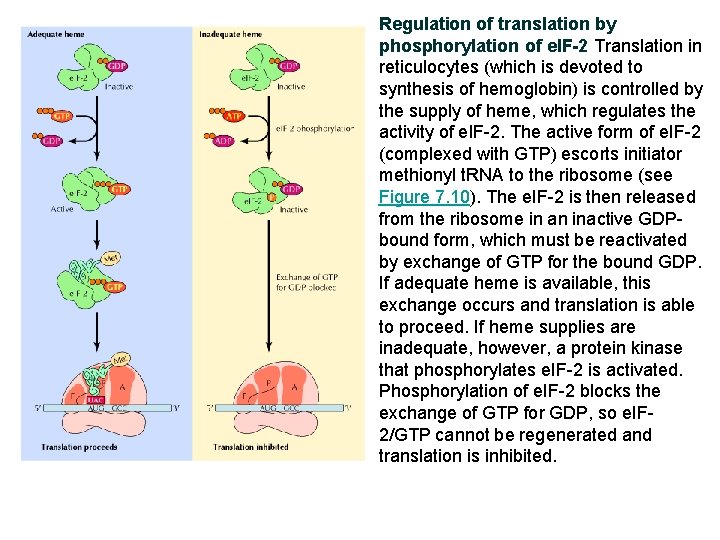
Regulation of translation by phosphorylation of e. IF-2 Translation in reticulocytes (which is devoted to synthesis of hemoglobin) is controlled by the supply of heme, which regulates the activity of e. IF-2. The active form of e. IF-2 (complexed with GTP) escorts initiator methionyl t. RNA to the ribosome (see Figure 7. 10). The e. IF-2 is then released from the ribosome in an inactive GDPbound form, which must be reactivated by exchange of GTP for the bound GDP. If adequate heme is available, this exchange occurs and translation is able to proceed. If heme supplies are inadequate, however, a protein kinase that phosphorylates e. IF-2 is activated. Phosphorylation of e. IF-2 blocks the exchange of GTP for GDP, so e. IF 2/GTP cannot be regenerated and translation is inhibited.

A possible reaction mechanism for the peptidyl transferase activity present in the large ribosomal subunit. The overall reaction is catalyzed by an active site in the 23 S r. RNA. In the first step of the proposed mechanism, the N 3 of the active-site adenine abstracts a proton from the amino acid attached to the t. RNA at the ribosome's Asite, allowing its amino nitrogen to attack the carboxyl group at the end of the growing peptide chain. In the next step this protonated adenine donates its hydrogen to the oxygen linked to the peptidyl-t. RNA, causing this t. RNA's release from the peptide chain. This leaves a polypeptide chain that is one amino acid longer than the starting reactants. The entire reaction cycle would then repeat with the next aminoacyl t. RNA that enters the A-site.

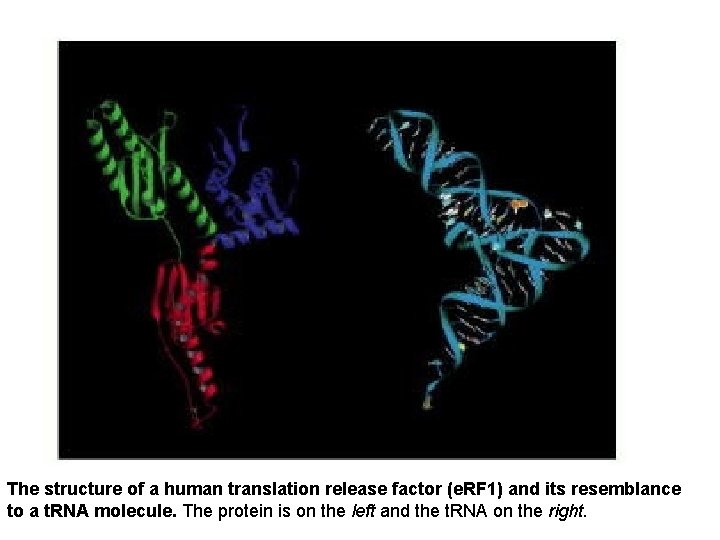
The structure of a human translation release factor (e. RF 1) and its resemblance to a t. RNA molecule. The protein is on the left and the t. RNA on the right.

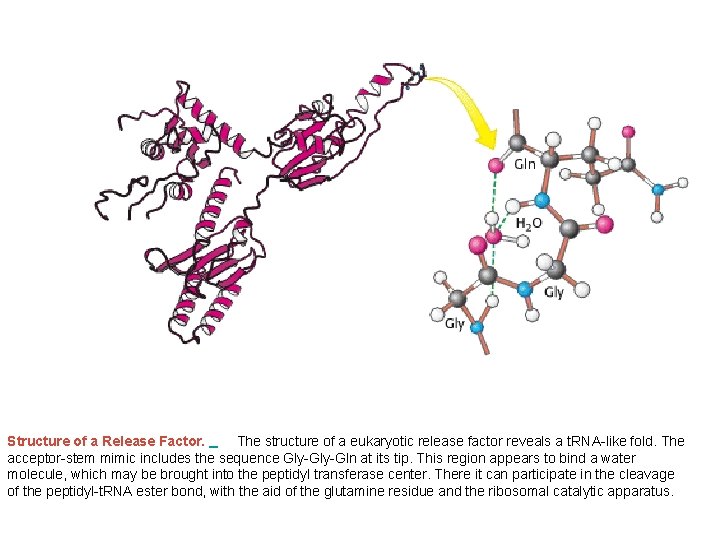
Structure of a Release Factor. The structure of a eukaryotic release factor reveals a t. RNA-like fold. The acceptor-stem mimic includes the sequence Gly-Gln at its tip. This region appears to bind a water molecule, which may be brought into the peptidyl transferase center. There it can participate in the cleavage of the peptidyl-t. RNA ester bond, with the aid of the glutamine residue and the ribosomal catalytic apparatus.

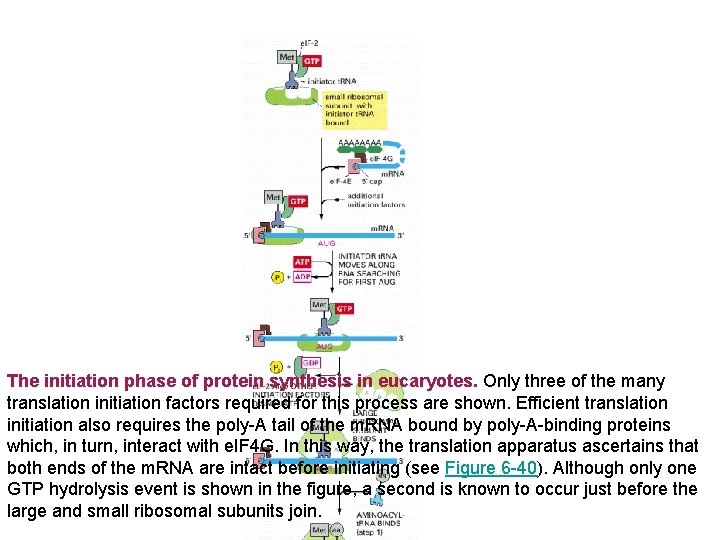
The initiation phase of protein synthesis in eucaryotes. Only three of the many translation initiation factors required for this process are shown. Efficient translation initiation also requires the poly-A tail of the m. RNA bound by poly-A-binding proteins which, in turn, interact with e. IF 4 G. In this way, the translation apparatus ascertains that both ends of the m. RNA are intact before initiating (see Figure 6 -40). Although only one GTP hydrolysis event is shown in the figure, a second is known to occur just before the large and small ribosomal subunits join.


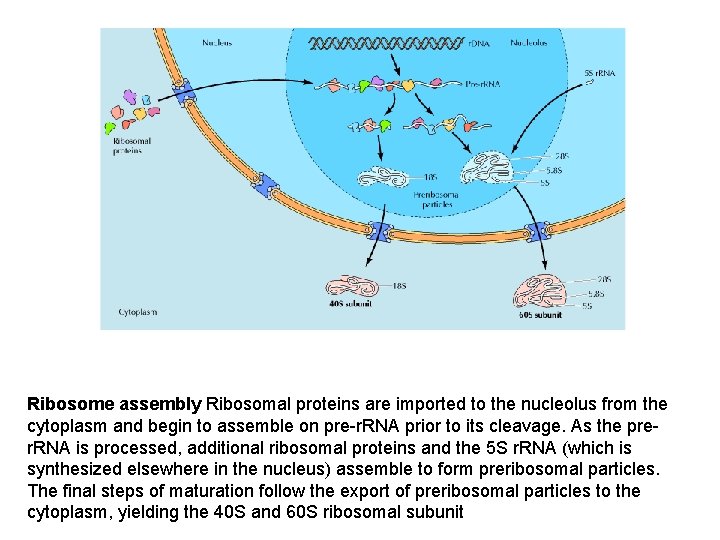
Ribosome assembly Ribosomal proteins are imported to the nucleolus from the cytoplasm and begin to assemble on pre-r. RNA prior to its cleavage. As the prer. RNA is processed, additional ribosomal proteins and the 5 S r. RNA (which is synthesized elsewhere in the nucleus) assemble to form preribosomal particles. The final steps of maturation follow the export of preribosomal particles to the cytoplasm, yielding the 40 S and 60 S ribosomal subunit

