Alternative splicing A playground of evolution Mikhail Gelfand












- Slides: 48

Alternative splicing: A playground of evolution Mikhail Gelfand Institute for Information Transmission Problems, RAS May 2004

Alternative splicing of human (and mouse) genes

• Alternative splicing of orthologous human and mouse genes • Sequence divergence in alternative and constitutive regions • Evolution of splicing sites • Alternative splicing and protein structure

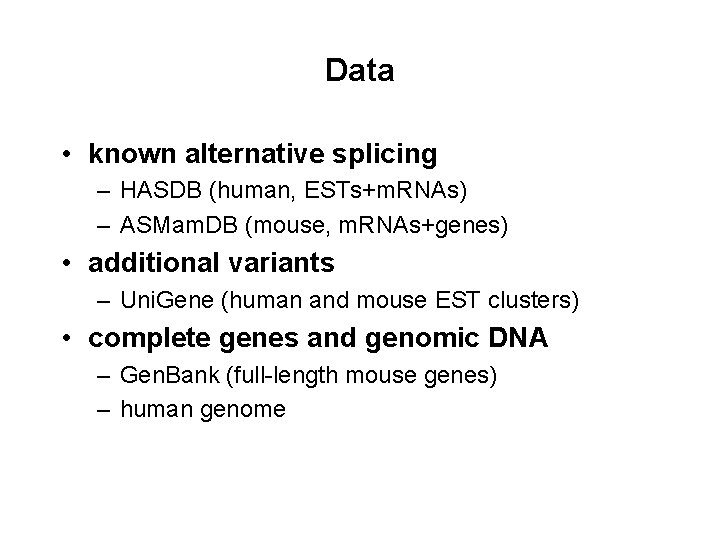
Data • known alternative splicing – HASDB (human, ESTs+m. RNAs) – ASMam. DB (mouse, m. RNAs+genes) • additional variants – Uni. Gene (human and mouse EST clusters) • complete genes and genomic DNA – Gen. Bank (full-length mouse genes) – human genome

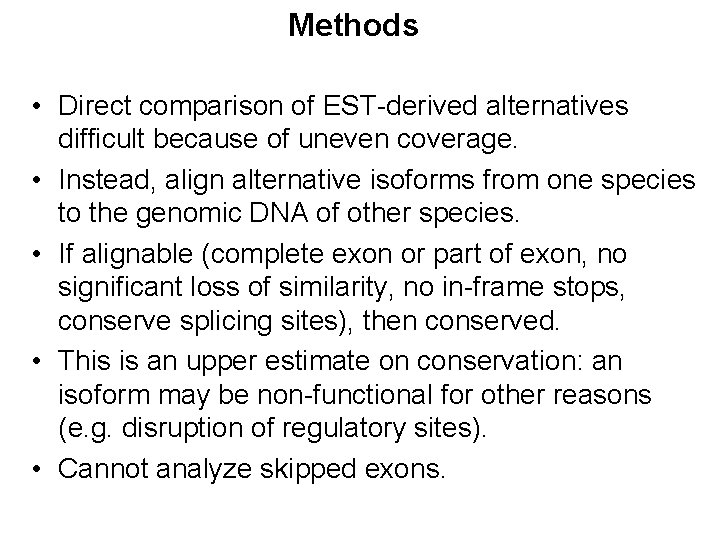
Methods • Direct comparison of EST-derived alternatives difficult because of uneven coverage. • Instead, align alternative isoforms from one species to the genomic DNA of other species. • If alignable (complete exon or part of exon, no significant loss of similarity, no in-frame stops, conserve splicing sites), then conserved. • This is an upper estimate on conservation: an isoform may be non-functional for other reasons (e. g. disruption of regulatory sites). • Cannot analyze skipped exons.

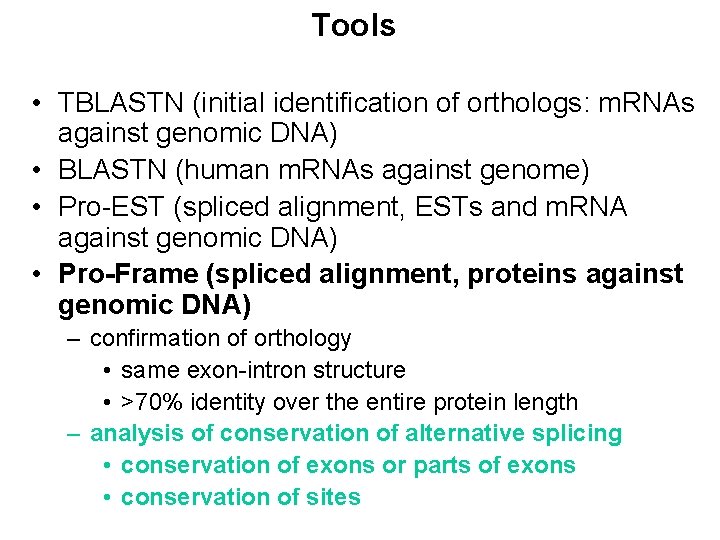
Tools • TBLASTN (initial identification of orthologs: m. RNAs against genomic DNA) • BLASTN (human m. RNAs against genome) • Pro-EST (spliced alignment, ESTs and m. RNA against genomic DNA) • Pro-Frame (spliced alignment, proteins against genomic DNA) – confirmation of orthology • same exon-intron structure • >70% identity over the entire protein length – analysis of conservation of alternative splicing • conservation of exons or parts of exons • conservation of sites

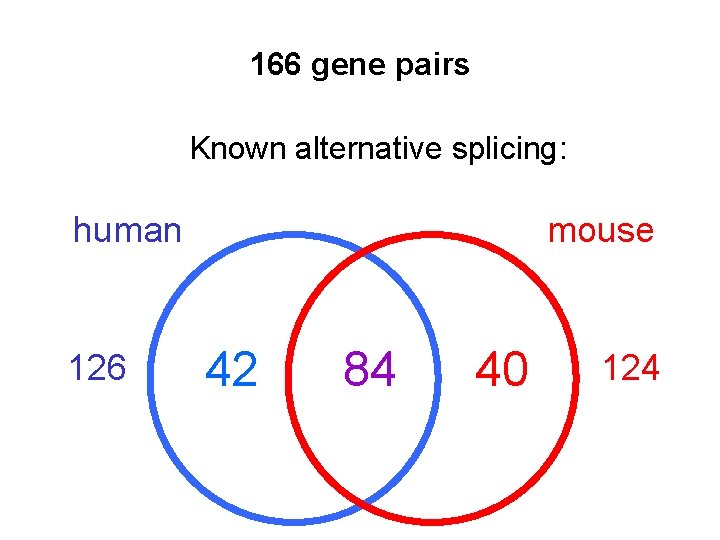
166 gene pairs Known alternative splicing: human 126 mouse 42 84 40 124

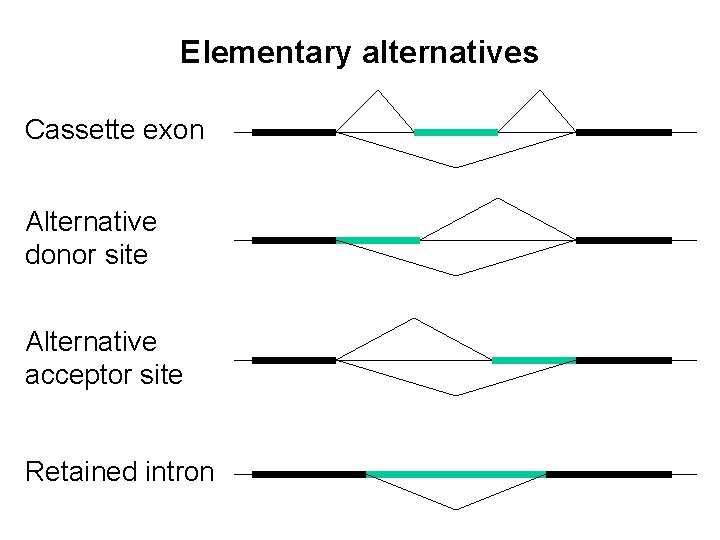
Elementary alternatives Cassette exon Alternative donor site Alternative acceptor site Retained intron

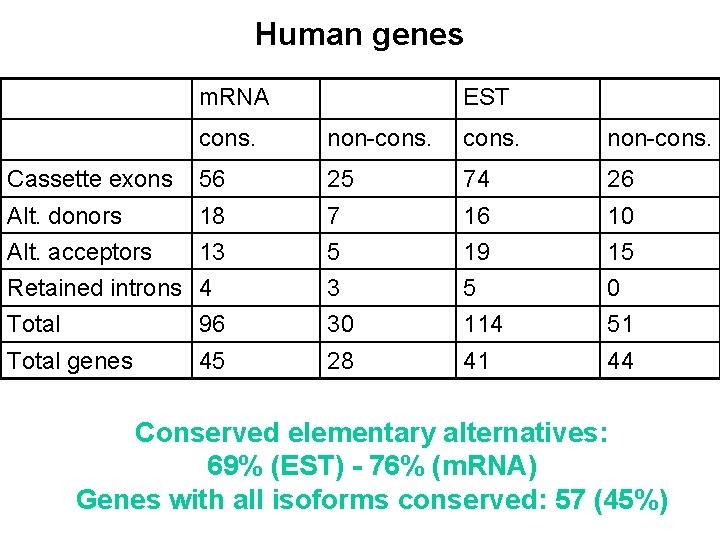
Human genes m. RNA EST cons. non-cons. Cassette exons 56 25 74 26 Alt. donors 18 7 16 10 Alt. acceptors 13 5 19 15 Retained introns 4 3 5 0 Total 96 30 114 51 Total genes 45 28 41 44 Conserved elementary alternatives: 69% (EST) - 76% (m. RNA) Genes with all isoforms conserved: 57 (45%)

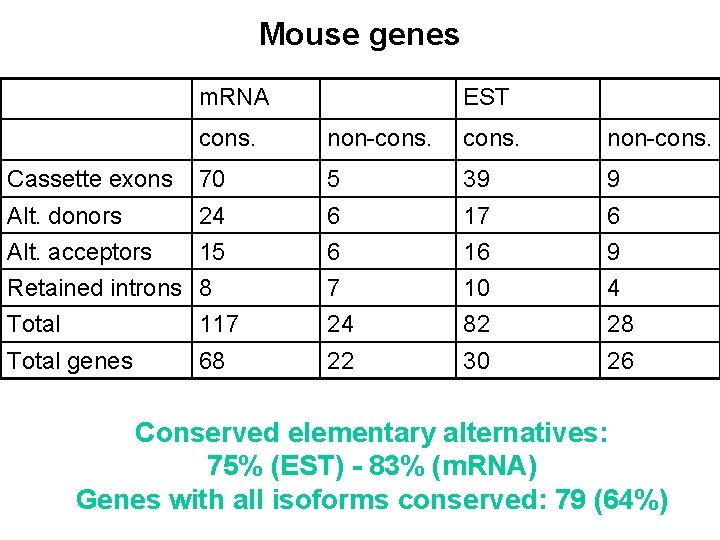
Mouse genes m. RNA EST cons. non-cons. Cassette exons 70 5 39 9 Alt. donors 24 6 17 6 Alt. acceptors 15 6 16 9 Retained introns 8 7 10 4 Total 117 24 82 28 Total genes 68 22 30 26 Conserved elementary alternatives: 75% (EST) - 83% (m. RNA) Genes with all isoforms conserved: 79 (64%)

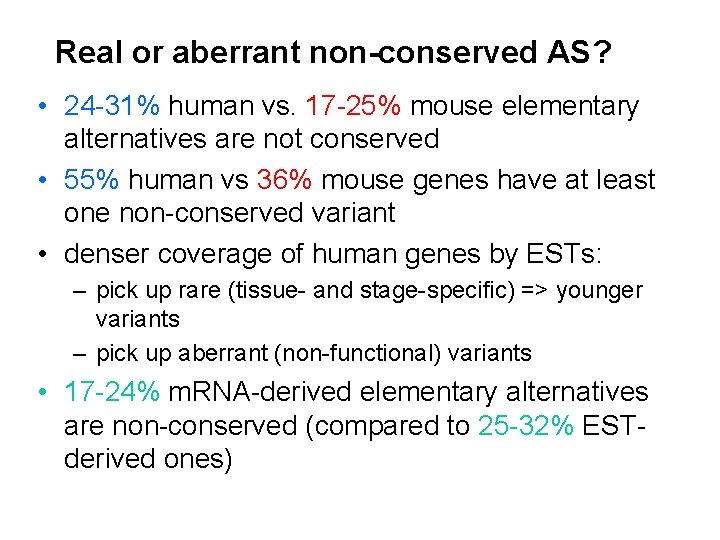
Real or aberrant non-conserved AS? • 24 -31% human vs. 17 -25% mouse elementary alternatives are not conserved • 55% human vs 36% mouse genes have at least one non-conserved variant • denser coverage of human genes by ESTs: – pick up rare (tissue- and stage-specific) => younger variants – pick up aberrant (non-functional) variants • 17 -24% m. RNA-derived elementary alternatives are non-conserved (compared to 25 -32% ESTderived ones)

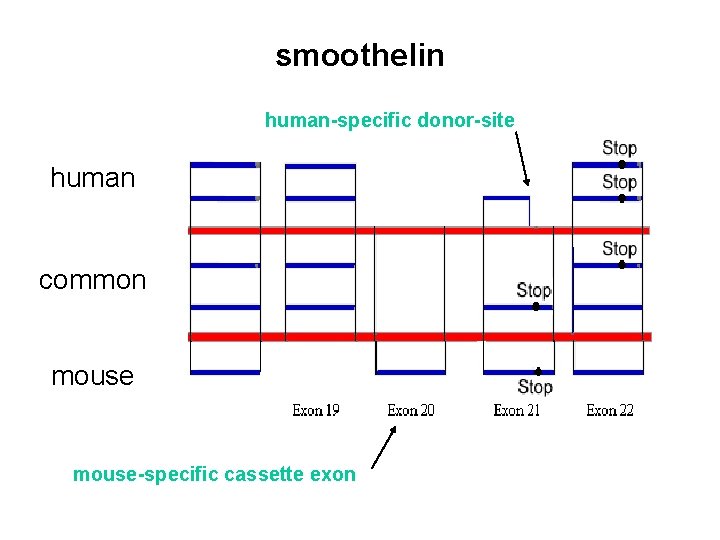
smoothelin human-specific donor-site human common mouse-specific cassette exon

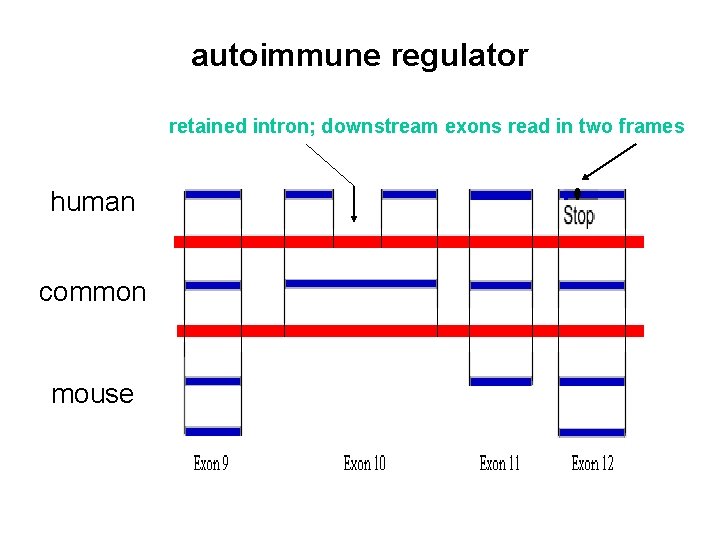
autoimmune regulator retained intron; downstream exons read in two frames human common mouse

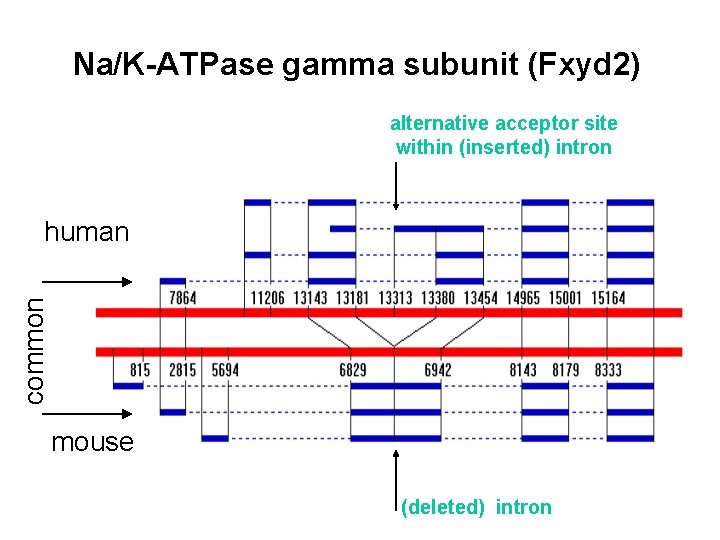
Na/K-ATPase gamma subunit (Fxyd 2) alternative acceptor site within (inserted) intron common human mouse (deleted) intron

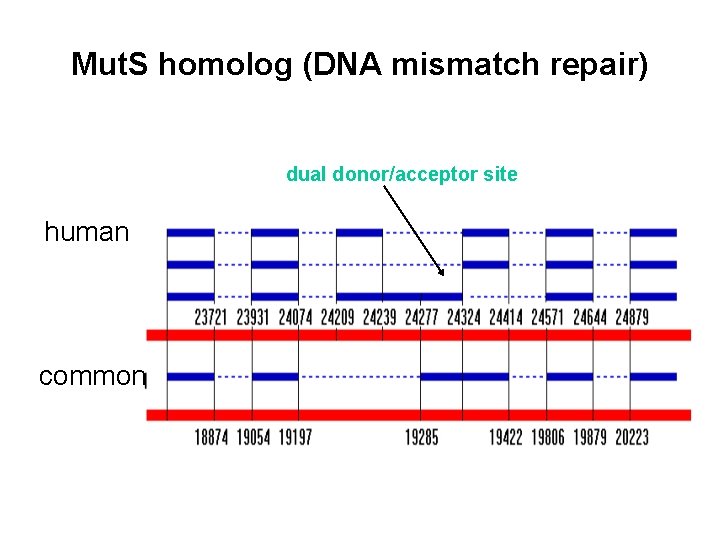
Mut. S homolog (DNA mismatch repair) dual donor/acceptor site human common

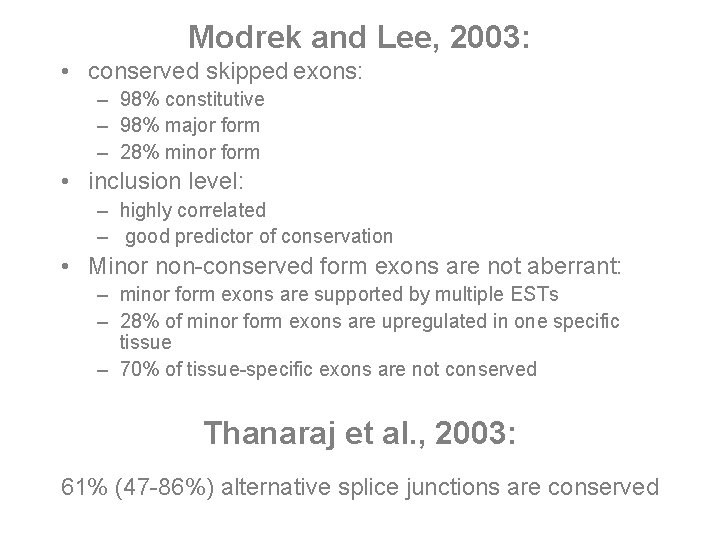
Modrek and Lee, 2003: • conserved skipped exons: – 98% constitutive – 98% major form – 28% minor form • inclusion level: – highly correlated – good predictor of conservation • Minor non-conserved form exons are not aberrant: – minor form exons are supported by multiple ESTs – 28% of minor form exons are upregulated in one specific tissue – 70% of tissue-specific exons are not conserved Thanaraj et al. , 2003: 61% (47 -86%) alternative splice junctions are conserved

• Alternative splicing of orthologous human and mouse genes • Sequence divergence in alternative and constitutive regions • Evolution of splicing sites • Alternative splicing and protein structure

“Contrary to our prediction, synonymous divergence between humans and non-human mammals was significantly higher in constitutive exons … Intriguingly, nonsynonymous divergence was marginally significantly higher in alternative exons” Iida and Akashi, 2000 Our preliminary observations: less synonymous, more non-synonymous divergence in alternative exons (human/mouse) => positive selection towards variability

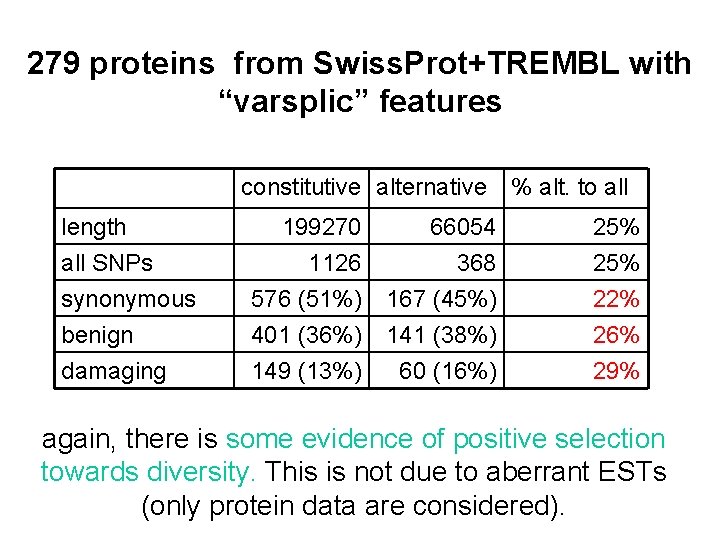
279 proteins from Swiss. Prot+TREMBL with “varsplic” features constitutive alternative % alt. to all length all SNPs synonymous 199270 1126 576 (51%) 66054 368 167 (45%) 25% 22% benign damaging 401 (36%) 149 (13%) 141 (38%) 60 (16%) 26% 29% again, there is some evidence of positive selection towards diversity. This is not due to aberrant ESTs (only protein data are considered).

• Alternative splicing of orthologous human and mouse genes • Sequence divergence in alternative and constitutive regions • Evolution of splicing sites • Alternative splicing and protein structure

Alternative splicing in a multigene family: the MAGEA family of cancer/testis specific antigens • A locus at the X chromosome containing eleven recently duplicated genes: two subfamilies of four genes each and three single genes • One protein-coding exon, multiple different 5’UTR exons • Originates from retroposed spliced m. RNA • Mutations create new splicing sites or disrupt existing sites

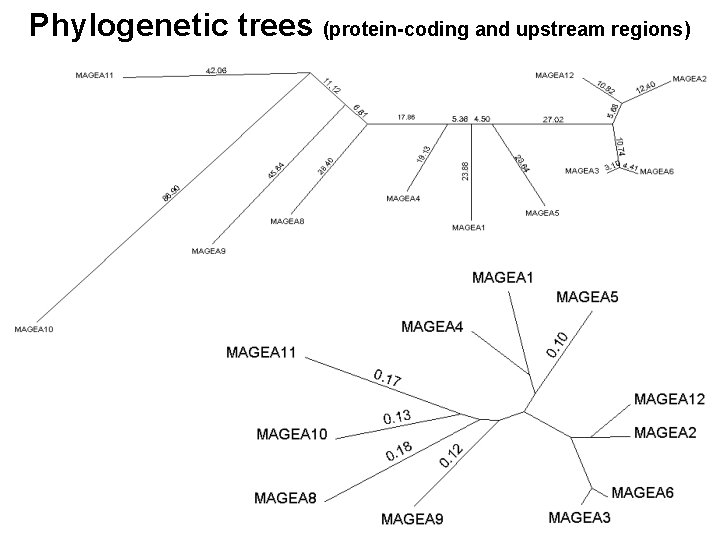
Phylogenetic trees (protein-coding and upstream regions)

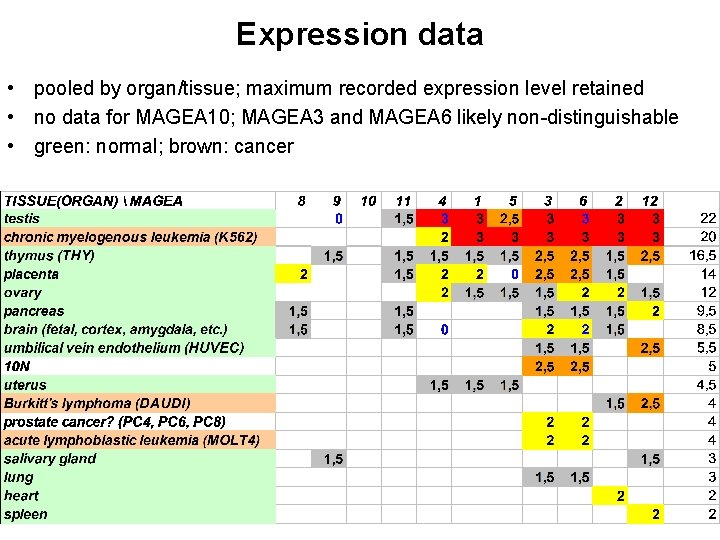
Expression data • pooled by organ/tissue; maximum recorded expression level retained • no data for MAGEA 10; MAGEA 3 and MAGEA 6 likely non-distinguishable • green: normal; brown: cancer

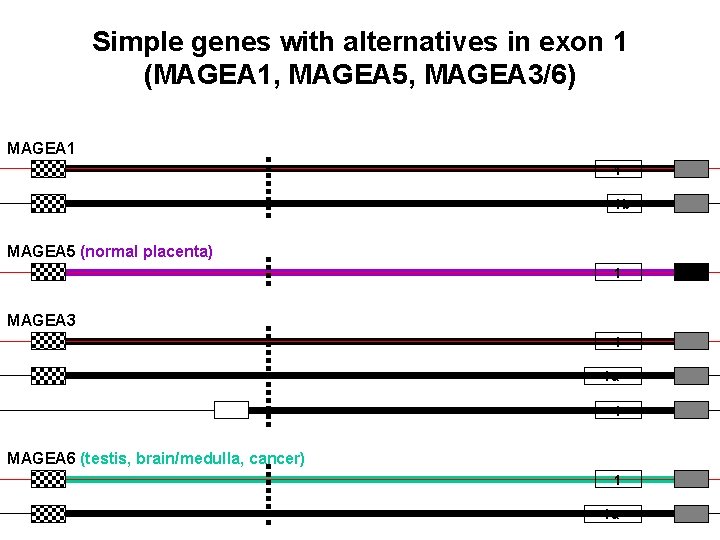
Simple genes with alternatives in exon 1 (MAGEA 1, MAGEA 5, MAGEA 3/6) MAGEA 1 1 1 b MAGEA 5 (normal placenta) 1 MAGEA 3 1 1 a 1 MAGEA 6 (testis, brain/medulla, cancer) 1 1 a

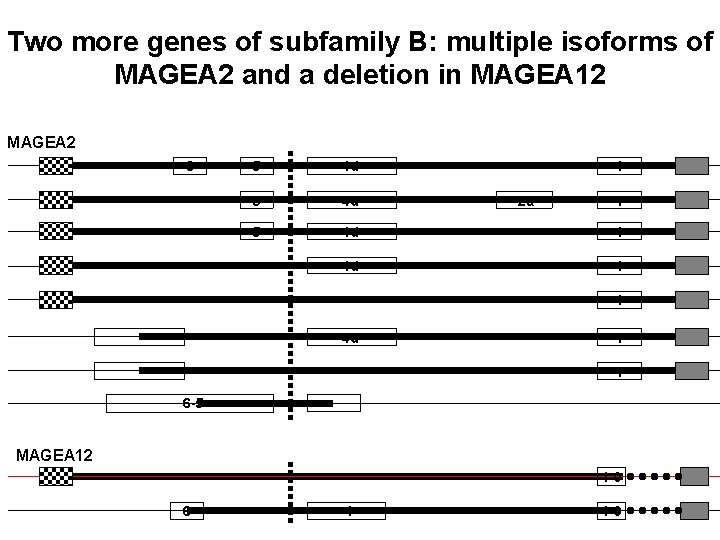
Two more genes of subfamily B: multiple isoforms of MAGEA 2 and a deletion in MAGEA 12 MAGEA 2 6 5 4 d 1 4 d 1 2 a 1 1 4 d 1 1 6 -5 MAGEA 12 1 -0 6 4 1 -0

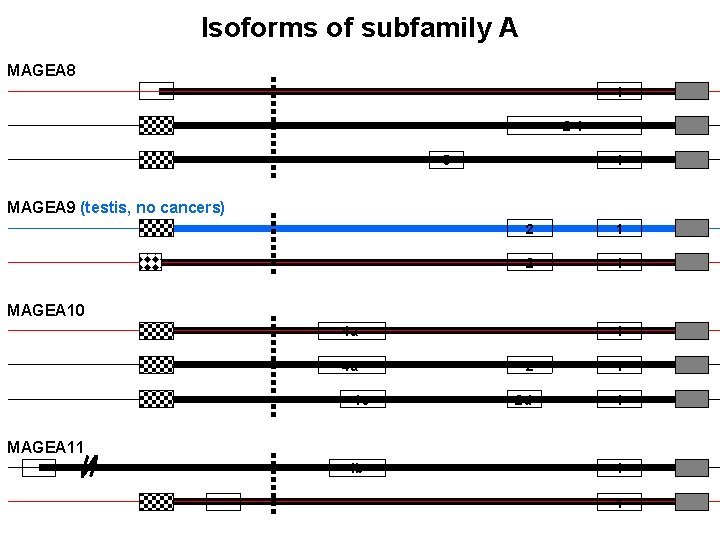
Isoforms of subfamily A MAGEA 8 1 2 -1 3 1 MAGEA 9 (testis, no cancers) 2 1 MAGEA 10 4 a 4 a 4 c 1 2 d 1 MAGEA 11 4 b 1 1

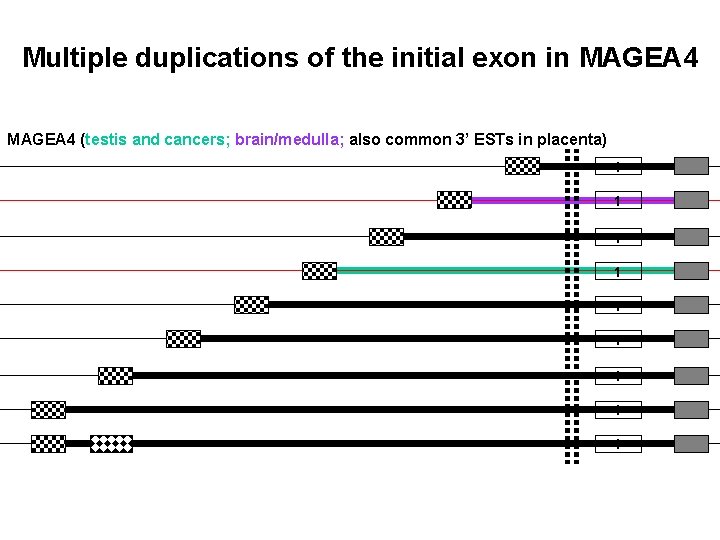
Multiple duplications of the initial exon in MAGEA 4 (testis and cancers; brain/medulla; also common 3’ ESTs in placenta) 1 1 1 1 1

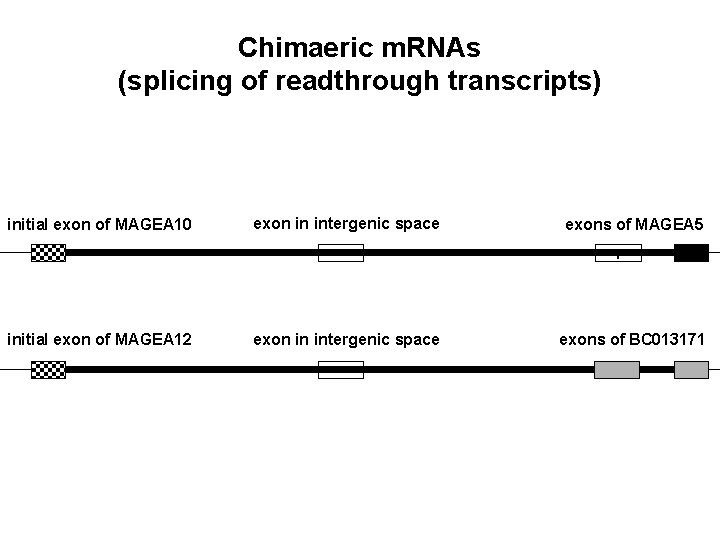
Chimaeric m. RNAs (splicing of readthrough transcripts) initial exon of MAGEA 10 exon in intergenic space exons of MAGEA 5 1 initial exon of MAGEA 12 exon in intergenic space exons of BC 013171

Other examples: • galactose-1 -phosphate uridylyltransferase + interleukin-11 receptor alpha chain (Magrangeas et al. , 1998) • P 2 Y 11 [receptor] + SSF 1 [nuclear protein] (Communi et al. , 2001) • Pr. P [Prion protein] + Dpl [prion-like protein Doppel] (Moore et al. , 1999) • cytochrome P 450 3 A: CYP 3 A 7 + two exons of a downstream pseudogene read in a different frame (Finta & Zaphiropoulos, 2000) • HHLA 1 + OC 90 [otoconin-90] (Kowalski et al. , 1999) • TRAX [translin-associated factor X] + DISC 1 [candidate schizophirenia gene] (Millar et al. , 2000) • Kua + UEV 1 [polyubiquination coeffector] (Thomson et al. , 2000) • FR + GAP [Rho GTPase activating protein] (Romani et al. , 2003) - ? • methyonyl t. RNA synthetase + advillin (Romani et al. , 2003) - ?

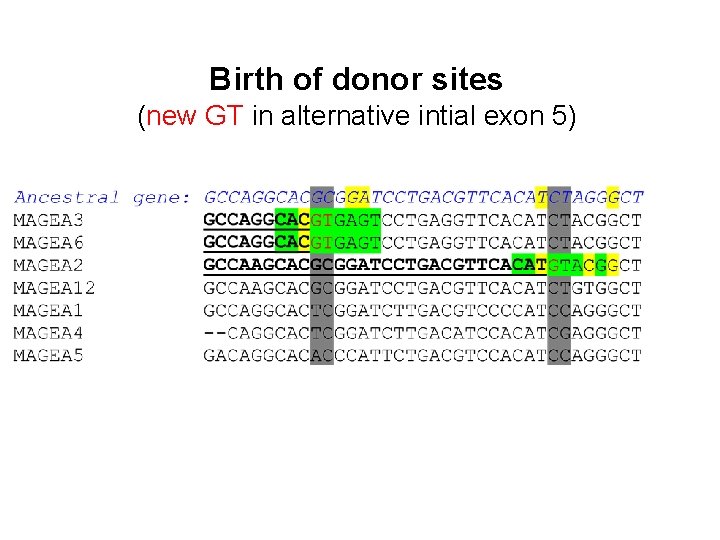
Birth of donor sites (new GT in alternative intial exon 5)

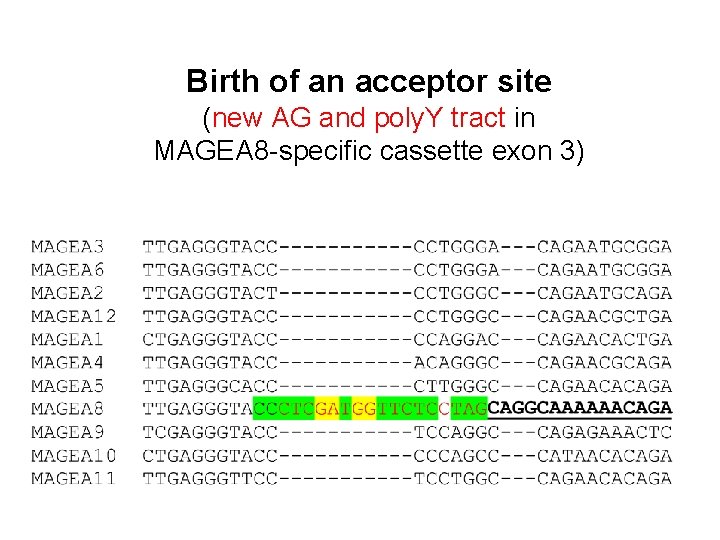
Birth of an acceptor site (new AG and poly. Y tract in MAGEA 8 -specific cassette exon 3)

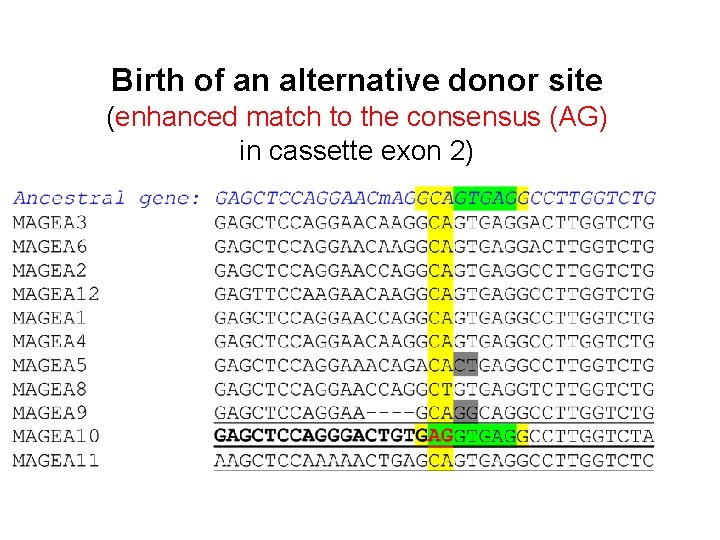
Birth of an alternative donor site (enhanced match to the consensus (AG) in cassette exon 2)

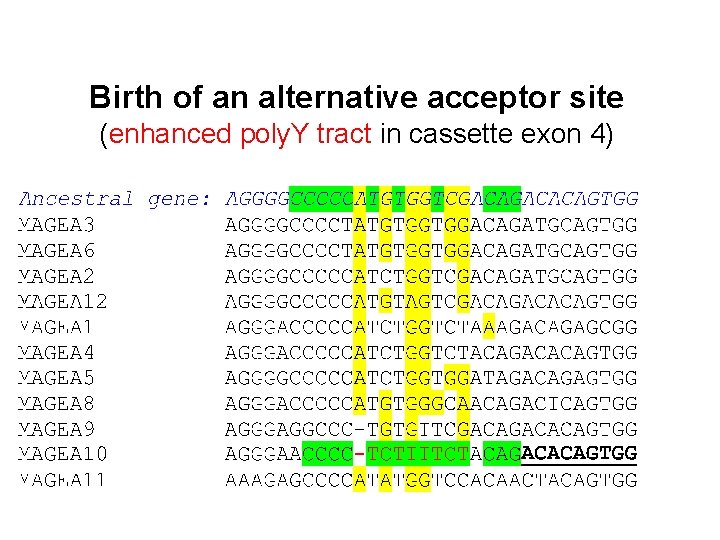
Birth of an alternative acceptor site (enhanced poly. Y tract in cassette exon 4)

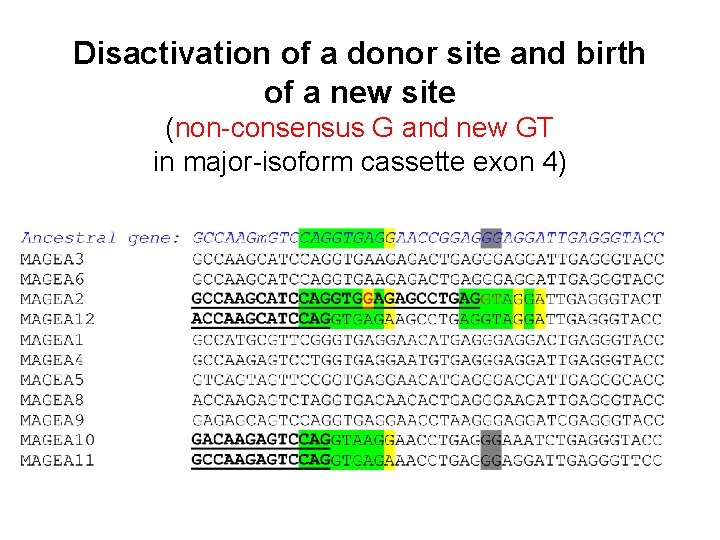
Disactivation of a donor site and birth of a new site (non-consensus G and new GT in major-isoform cassette exon 4)

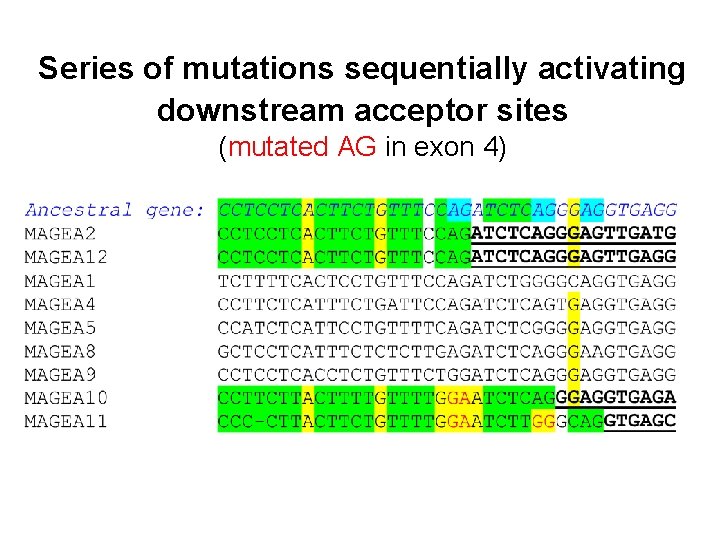
Series of mutations sequentially activating downstream acceptor sites (mutated AG in exon 4)

• Alternative splicing of orthologous human and mouse genes • Sequence divergence in alternative and constitutive regions • Evolution of splicing sites • Alternative splicing and protein structure

Data • Alternatively spliced genes (proteins) from Swiss. Prot – human – mouse • Protein structures from PDB • Domains from Inter. Pro – – SMART Pfam Prosite etc.

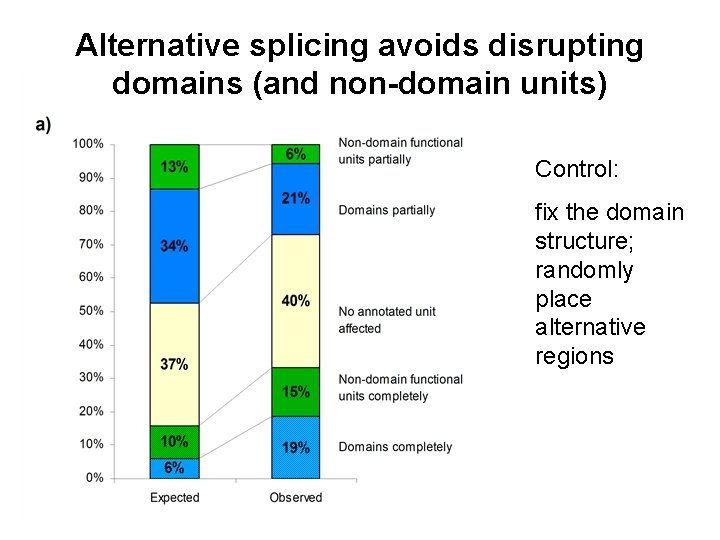
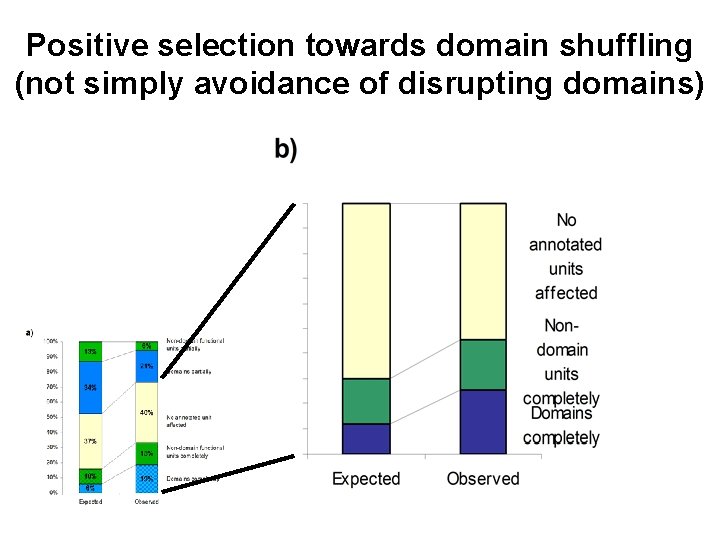
Alternative splicing avoids disrupting domains (and non-domain units) Control: fix the domain structure; randomly place alternative regions

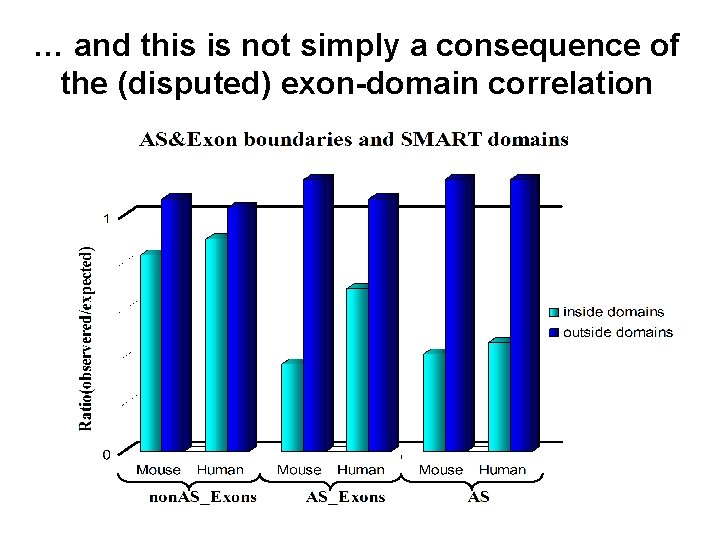
… and this is not simply a consequence of the (disputed) exon-domain correlation

Positive selection towards domain shuffling (not simply avoidance of disrupting domains)

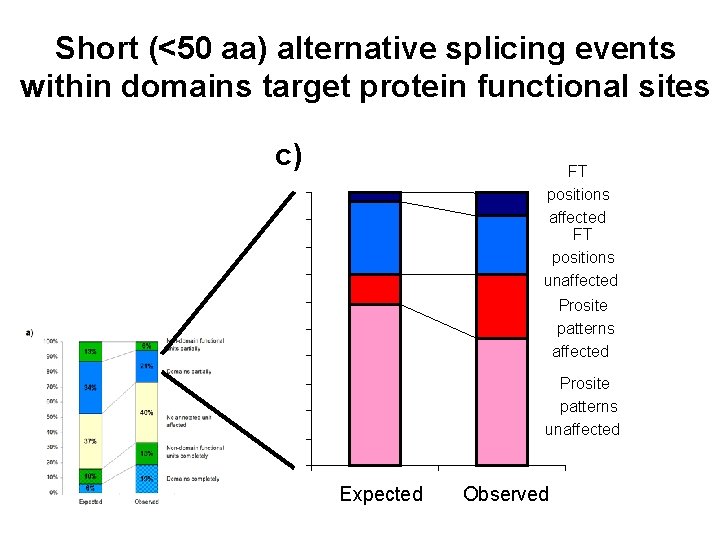
Short (<50 aa) alternative splicing events within domains target protein functional sites c) FT positions affected FT positions unaffected Prosite patterns unaffected Expected Observed

An attempt of integration • AS is often young (as opposed to degenerating) • young AS isoforms are often minor and tissuespecific • … but still functional – although unique isoforms may be result of aberrant splicing • AS regions show evidence for positive selection – excess damaging SNPs – excess non-synonymous codon substitutions • MAGEA - not aberrant, because explainable by effects of mutations

What to do • Each isoform (alternative region) can be characterized: – by conservation (between genomes) – if conserved, by selection (positive vs negative) • human-mouse, also add rat – pattern of SNPs (synonymous, benign, damaging) – tissue-specificity • in particular, whether it is cancer-specific – degree of inclusion (major/minor) – functionality (for isoforms) • whether it generates a frameshift • how bad it is (the distance between the stop-codon and the last exon-exon junction)

What to expect (hypotheses) • Cancer-specific isoforms will be less functional and more often non-conserved • Non-conserved isoforms will contain a larger fraction of non-functional isoforms; and this may influence evolutionary conclusions • Still, after removal of non-functional isoforms, one should see positive selection in alternative regions (more non-synonymous substitutions compared to constant regions etc. ); especially in tissue-specific ones.

Plans • careful and detailed analysis of human-mouse -(rat)-((dog)) AS isoforms (human and mouse ESTs) • conservation of AS regulatory sites • mosquito-drosophila • more families of paralogs; add mouse data • AS of transcription factors and receptors

Acknowledgements • Discussions – – Vsevolod Makeev (Gos. NIIGenetika) Eugene Koonin (NCBI) Igor Rogozin (NCBI) Dmitry Petrov (Stanford) • Support – Ludwig Institute of Cancer Research – Howard Hughes Medical Institute

Authors • Andrei Mironov (Gos. NIIGenetika) – spliced alignment • Shamil Sunyaev (EMBL, now Harvard University Medical School) – protein structure • Vasily Ramensky (Institute of Molecular Biology) – SNPs • Irena Artamonova (Institute of Bioorganic Chemistry) – human/mouse comparison, MAGEA family • Dmitry Malko (Gos. NIIGenetika) – mosquito/drosophila comparison • Eugenia Kriventseva (EBI, now BASF) – protein structure • Ramil Nurtdinov (Moscow State University) – human/mouse comparison • Ekaterina Ermakova (Moscow State University) – evolution of alternative/constitutive regions

References Nurtdinov RN, Artamonova II, Mironov AA, Gelfand MS (2003) Low conservation of alternative splicing patterns in the human and mouse genomes. Human Molecular Genetics 12: 1313 -1320. Kriventseva EV, Koch I, Apweiler R, Vingron M, Bork P, Gelfand MS, Sunyaev S. (2003) Increase of functional diversity by alternative splicing. Trends in Genetics 19: 124 -128. Brudno M, Gelfand MS, Spengler S, Zorn M, Dubchak I, Conboy JG (2001) Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-m. RNA splicing. Nucleic Acids Research 29: 2338 -2348. Dralyuk I, Brudno M, Gelfand MS, Zorn M, Dubchak I (2000) ASDB: database of alternatively spliced genes. Nucleic Acids Research 28: 296 -297. Mironov AA, Fickett JW, Gelfand MS (1999). Frequent alternative splicing of human genes. Genome Research 9: 1288 -1293.