Algorithms Lecture 11 String matching problems Longest common
Algorithms Lecture 11
“String matching” problems • Longest common subsequence • RNA secondary structure • Sequence alignment • Motivation: – Additional examples of dynamic programming – Applications

Longest common subsequence • Given a string X=x 1 x 2…xm over some alphabet, X’ is a subsequence of X if there are i 1 < … < ik such that X’ = xi 1 xi 2…xik – E. g. , “sven” is a subsequence of “seventeen” • Given two strings X, Y, a longest common subsequence is a string Z of longest length that is a subsequence of both X and Y – Need not be unique

Algorithms for LCS? • Brute-force solution? – Number of subsequences of a given string is exponential in the length of the string • Try dynamic programming!
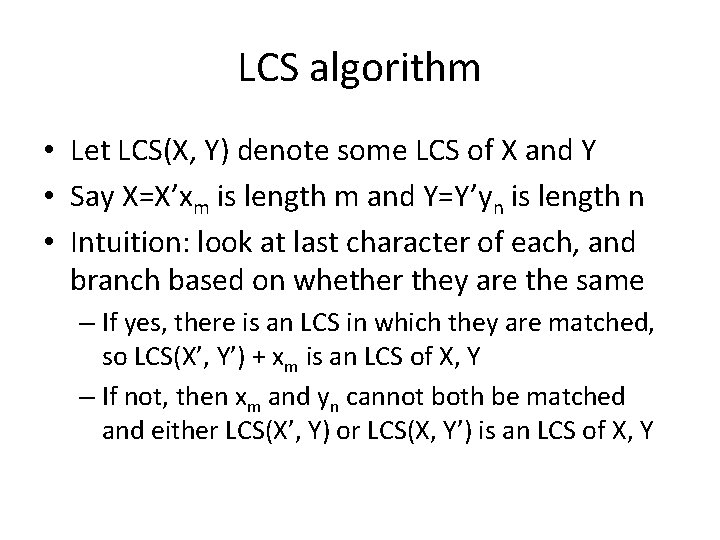
LCS algorithm • Let LCS(X, Y) denote some LCS of X and Y • Say X=X’xm is length m and Y=Y’yn is length n • Intuition: look at last character of each, and branch based on whether they are the same – If yes, there is an LCS in which they are matched, so LCS(X’, Y’) + xm is an LCS of X, Y – If not, then xm and yn cannot both be matched and either LCS(X’, Y) or LCS(X, Y’) is an LCS of X, Y
![LCS algorithm • Store 2 D array c where c[i, j] is the length LCS algorithm • Store 2 D array c where c[i, j] is the length](http://slidetodoc.com/presentation_image_h2/8fd7184fc7694e0fe2981a36dac6f798/image-6.jpg)
LCS algorithm • Store 2 D array c where c[i, j] is the length of the LCS of x 1…xi and y 1…yj – The length of the LCS of X, Y is c[m, n] • Base case: c[i, 0] = c[0, j] = 0 • For i, j > 0: c[i, j] = 1 + c[i-1, j-1] if xi = yj c[i, j] = max{ c[i, j-1], c[i-1, j] } if xi yj

LCS algorithm • Can implement recursively or (better) iteratively, as we have seen before • Running time easiest to analyze for iterative algorithm: – O(mn) entries in the array, each can be computed in constant time (given previous entries) O(mn) running time overall • Algorithm can be extended to return the LCS itself
RNA secondary structure • RNA is a single-stranded molecule consisting of a linear sequence of bases {A, C, G, U} • RNA molecule “folds back on itself” if different bases are aligned with each other – A-U or C-G • Most-stable configuration is one in which the maximum number of bases are aligned (subject to some additional rules) • This is called “secondary structure” of RNA – Determining secondary structure is an important problem in computational biology
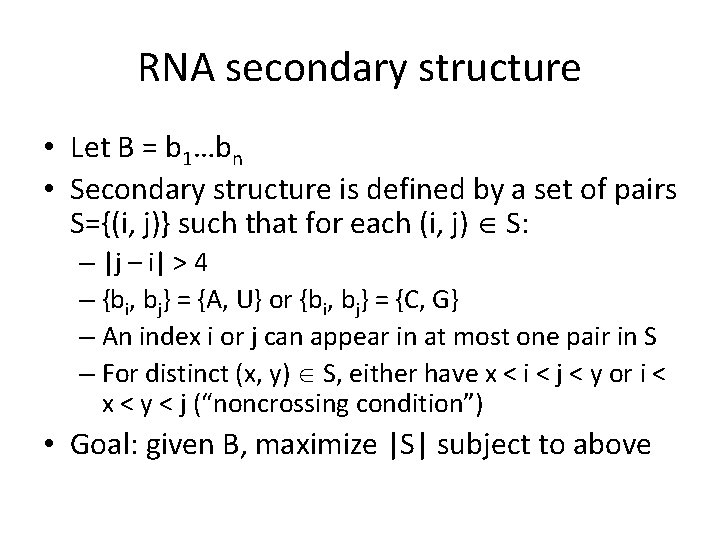
RNA secondary structure • Let B = b 1…bn • Secondary structure is defined by a set of pairs S={(i, j)} such that for each (i, j) S: – |j – i| > 4 – {bi, bj} = {A, U} or {bi, bj} = {C, G} – An index i or j can appear in at most one pair in S – For distinct (x, y) S, either have x < i < j < y or i < x < y < j (“noncrossing condition”) • Goal: given B, maximize |S| subject to above

Dynamic-programming solution • First attempt: let Opt(j) be the size of the largest set for the prefix b 1…bj • To compute Opt(j): – Either bj is aligned with another base in an optimal solution, or not – If not, then Opt(j) = Opt(j-1) – Else, bj is aligned with bt for some t < j-4 • Opt(j) = 1 + Opt(t-1) + ? ? • Need to store Opt for all contiguous substrings
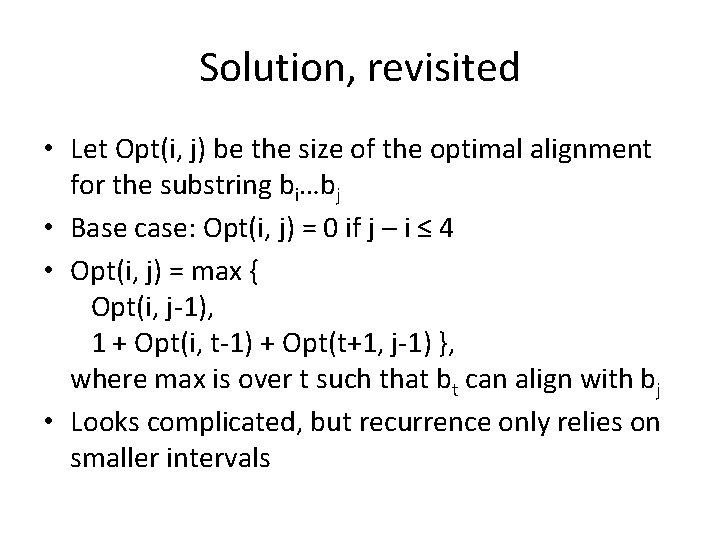
Solution, revisited • Let Opt(i, j) be the size of the optimal alignment for the substring bi…bj • Base case: Opt(i, j) = 0 if j – i ≤ 4 • Opt(i, j) = max { Opt(i, j-1), 1 + Opt(i, t-1) + Opt(t+1, j-1) }, where max is over t such that bt can align with bj • Looks complicated, but recurrence only relies on smaller intervals

Running time? • O(n 2) array entries to fill… – …each takes time O(n) to compute O(n 3) running time overall
- Slides: 12