Alberts Johnson Lewis Raff Roberts Walter Molecular Biology
Alberts • Johnson • Lewis • Raff • Roberts • Walter Molecular Biology of the Cell Fifth Edition Chapter 6 How Cells Read the Genome: From DNA to Protein Copyright © Garland Science 2008
Chapter 6 • How Cells Read the Genome: From DNA to Protein • • I) RNA II) Eubacterial transcription III) Eubacterial Initiation and Termination IV) Eukaryotic polymerases V) Eukaryotic Initiation VI Elongation VII Post-transcriptional modification (capping, splicing, cleavage and poly adenylation).
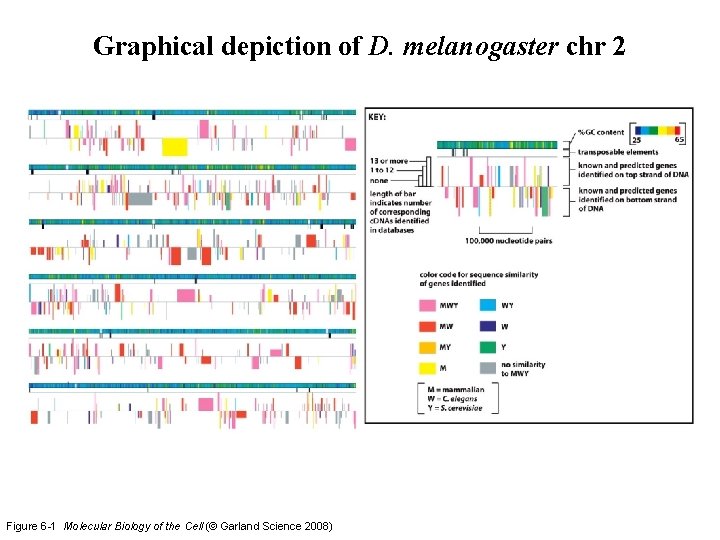
Graphical depiction of D. melanogaster chr 2 Figure 6 -1 Molecular Biology of the Cell (© Garland Science 2008)
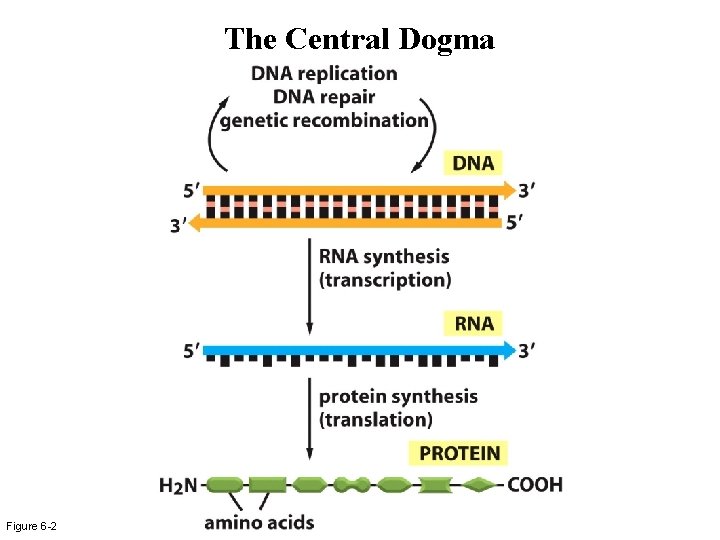
The Central Dogma Figure 6 -2
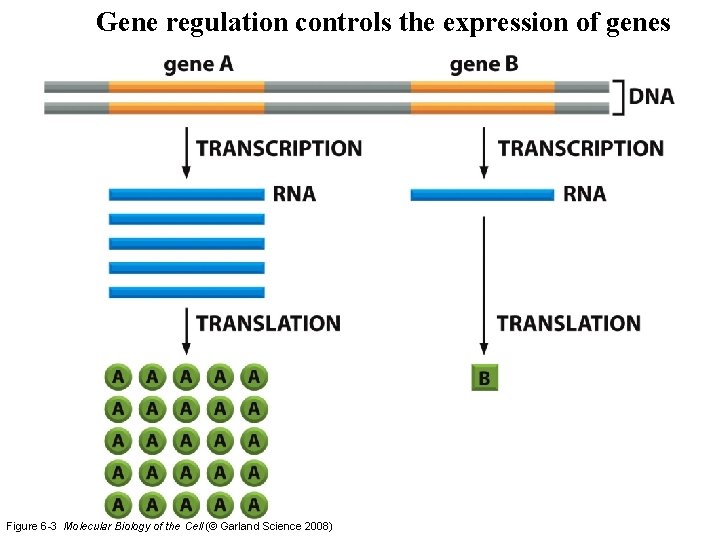
Gene regulation controls the expression of genes Figure 6 -3 Molecular Biology of the Cell (© Garland Science 2008)
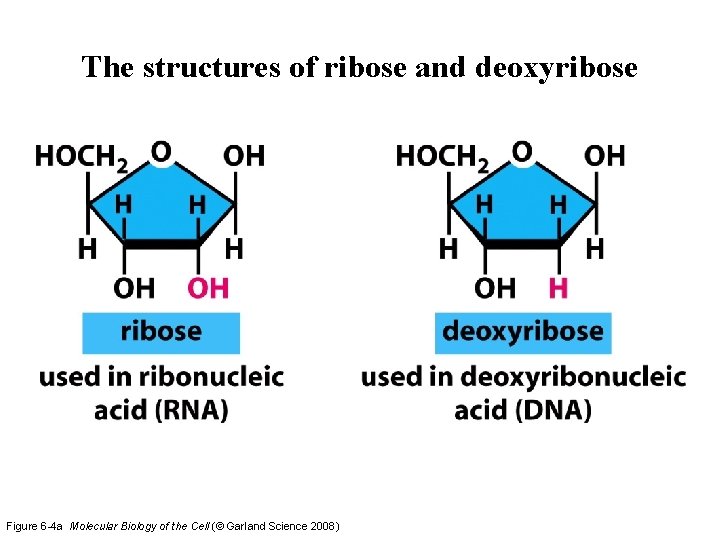
The structures of ribose and deoxyribose Figure 6 -4 a Molecular Biology of the Cell (© Garland Science 2008)
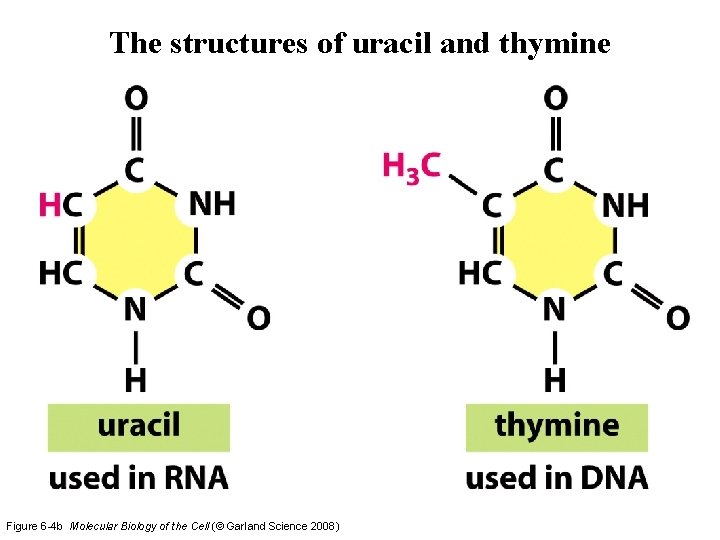
The structures of uracil and thymine Figure 6 -4 b Molecular Biology of the Cell (© Garland Science 2008)
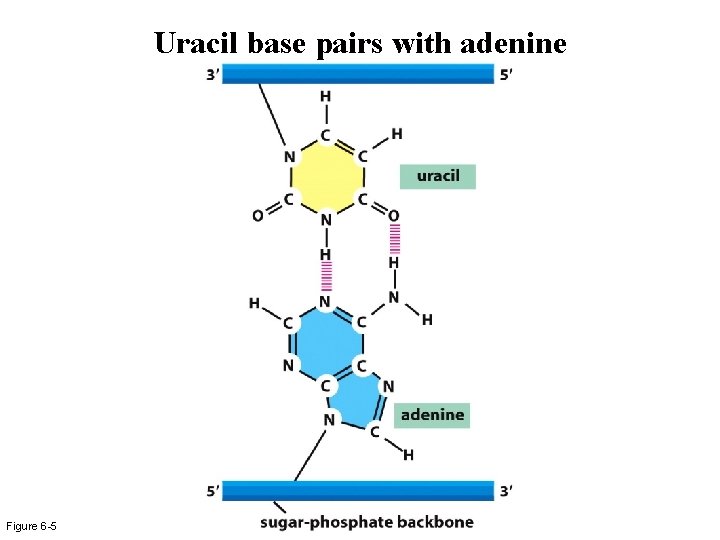
Uracil base pairs with adenine Figure 6 -5
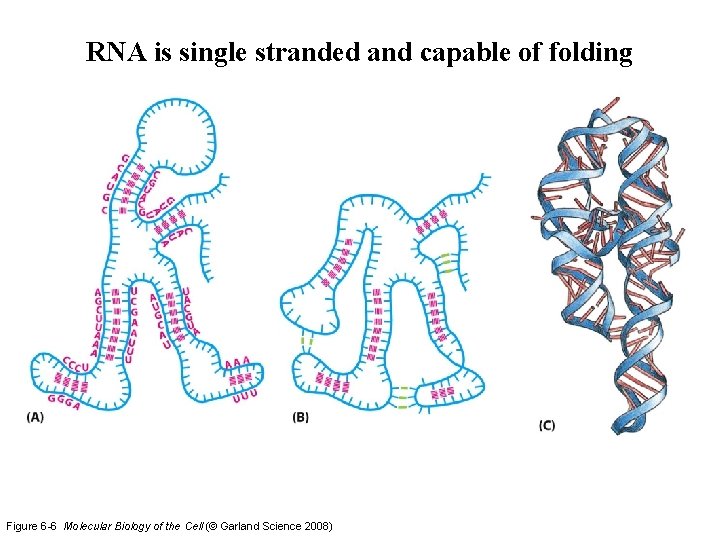
RNA is single stranded and capable of folding Figure 6 -6 Molecular Biology of the Cell (© Garland Science 2008)
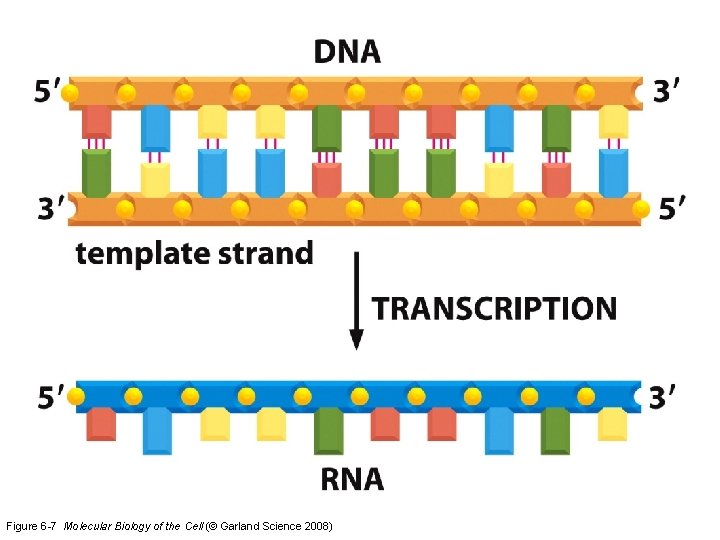
Figure 6 -7 Molecular Biology of the Cell (© Garland Science 2008)
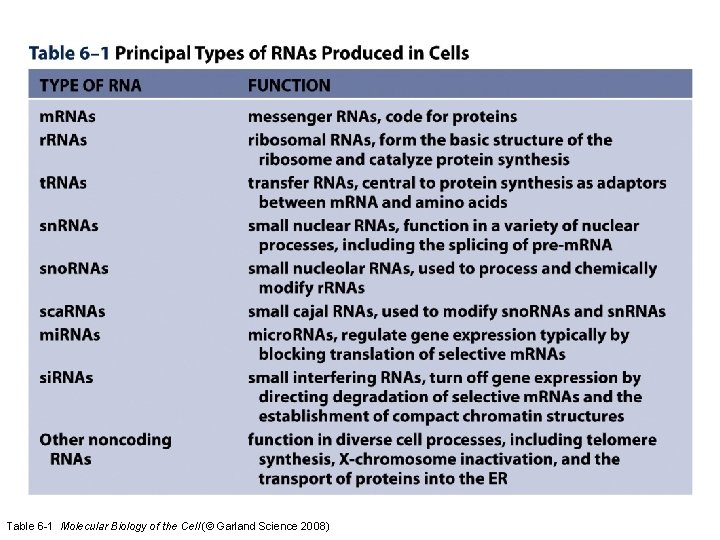
Table 6 -1 Molecular Biology of the Cell (© Garland Science 2008)
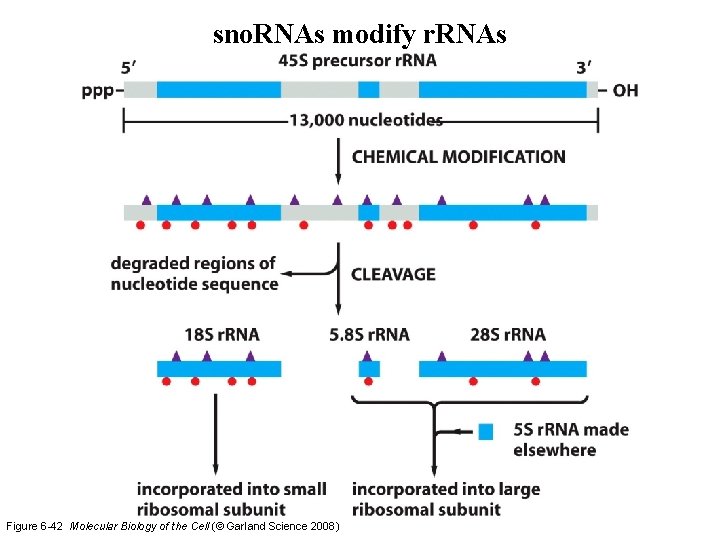
sno. RNAs modify r. RNAs Figure 6 -42 Molecular Biology of the Cell (© Garland Science 2008)
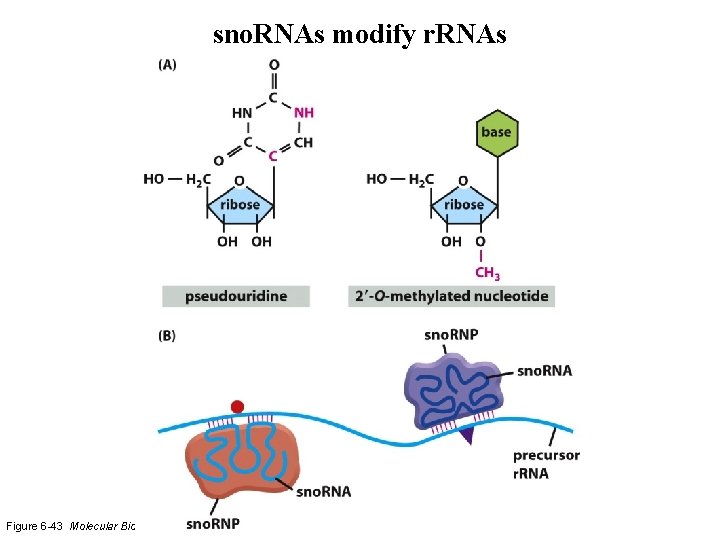
sno. RNAs modify r. RNAs Figure 6 -43 Molecular Biology of the Cell (© Garland Science 2008)
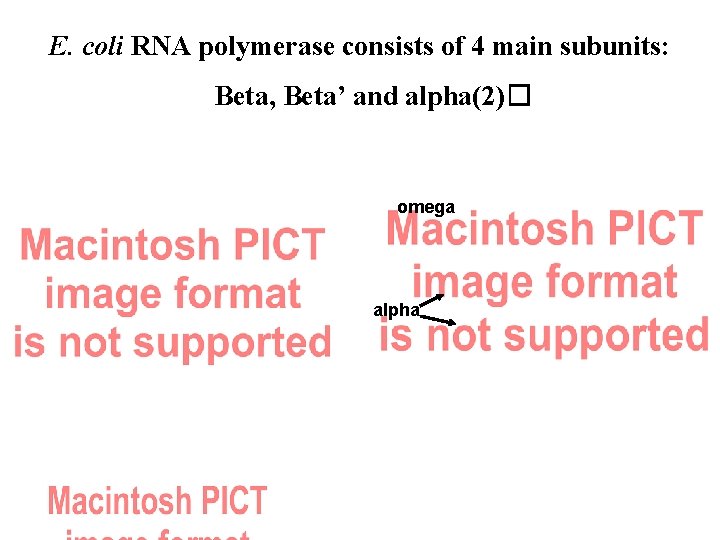
E. coli RNA polymerase consists of 4 main subunits: Beta, Beta’ and alpha(2)� omega alpha
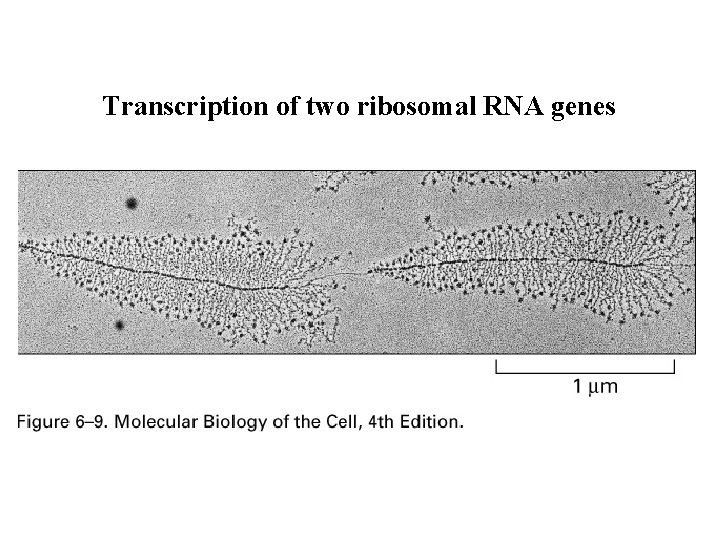
Transcription of two ribosomal RNA genes
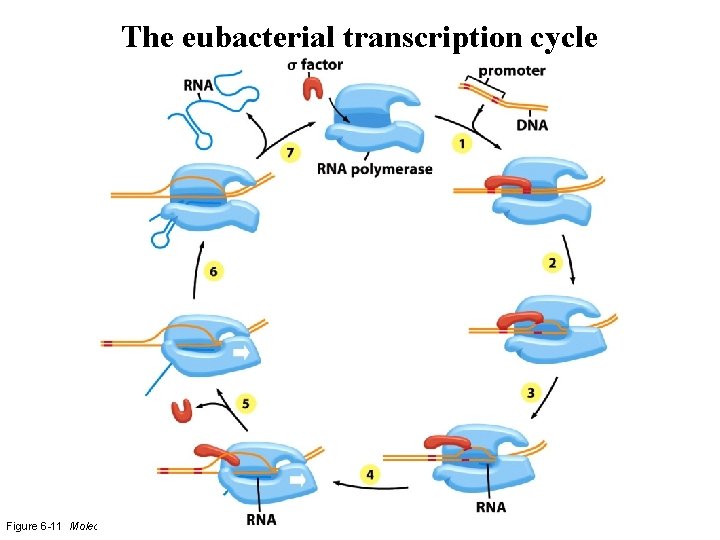
The eubacterial transcription cycle Figure 6 -11 Molecular Biology of the Cell (© Garland Science 2008)
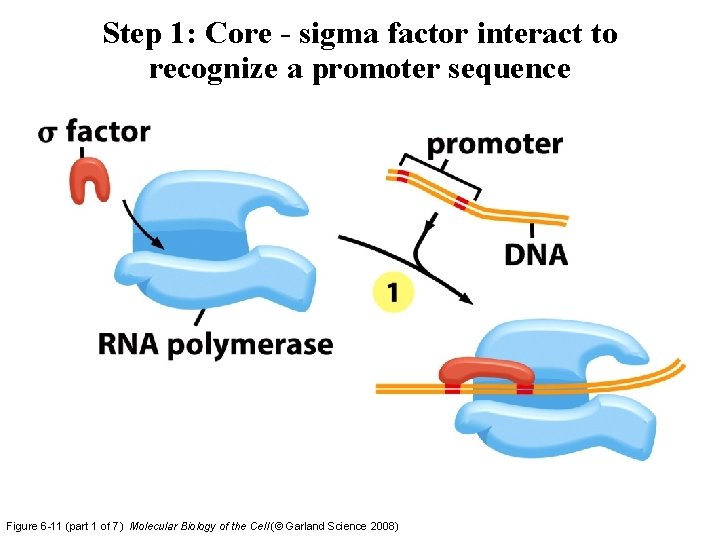
Step 1: Core - sigma factor interact to recognize a promoter sequence Figure 6 -11 (part 1 of 7) Molecular Biology of the Cell (© Garland Science 2008)
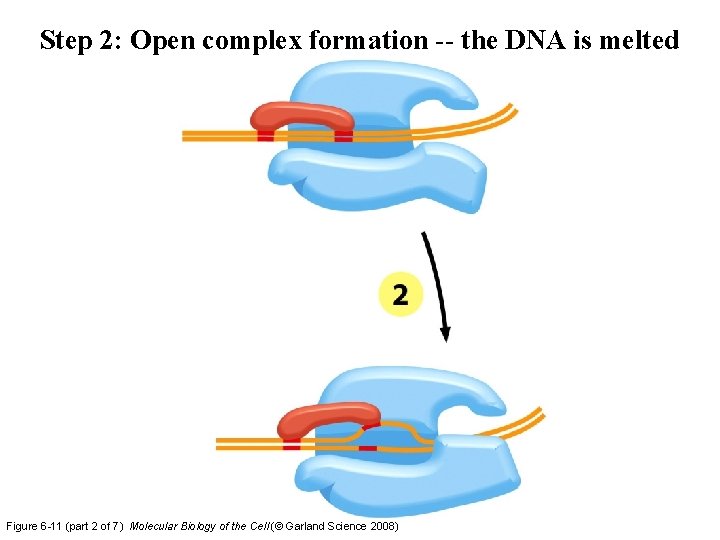
Step 2: Open complex formation -- the DNA is melted Figure 6 -11 (part 2 of 7) Molecular Biology of the Cell (© Garland Science 2008)
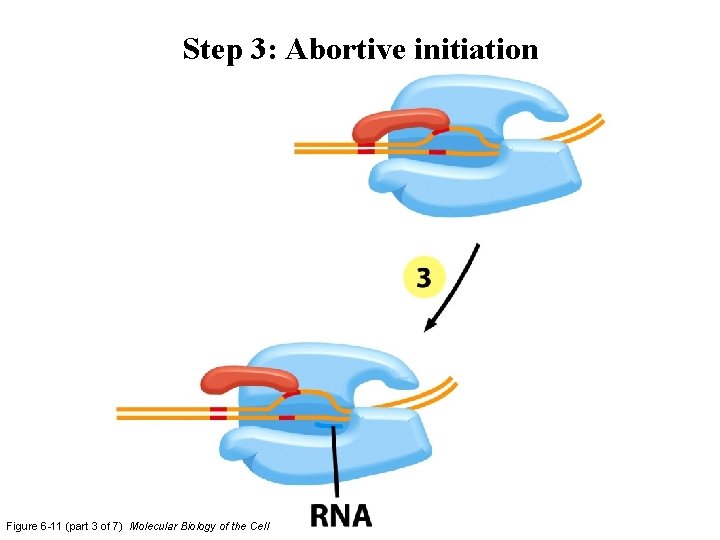
Step 3: Abortive initiation Figure 6 -11 (part 3 of 7) Molecular Biology of the Cell
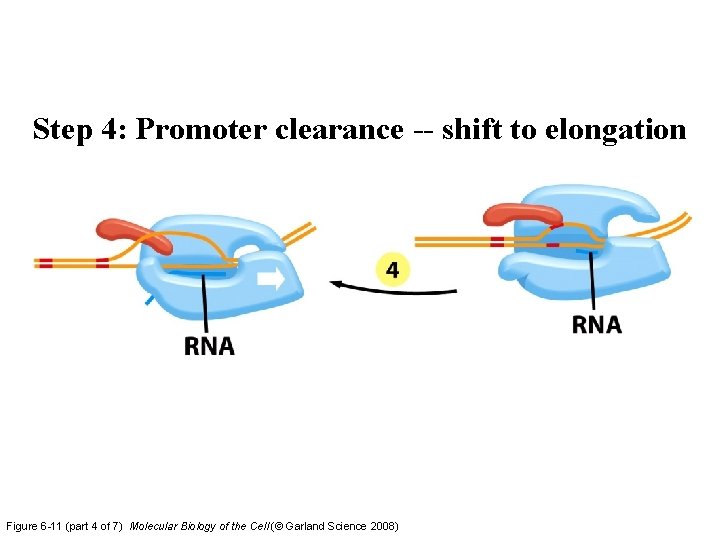
Step 4: Promoter clearance -- shift to elongation Figure 6 -11 (part 4 of 7) Molecular Biology of the Cell (© Garland Science 2008)
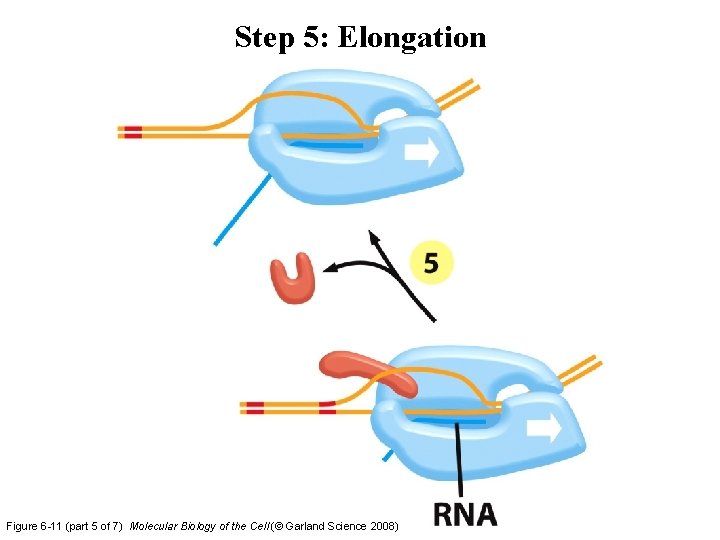
Step 5: Elongation Figure 6 -11 (part 5 of 7) Molecular Biology of the Cell (© Garland Science 2008)
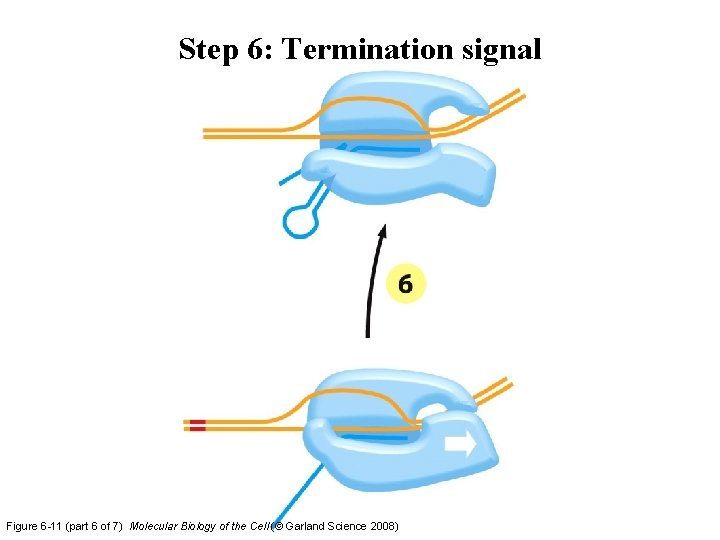
Step 6: Termination signal Figure 6 -11 (part 6 of 7) Molecular Biology of the Cell (© Garland Science 2008)
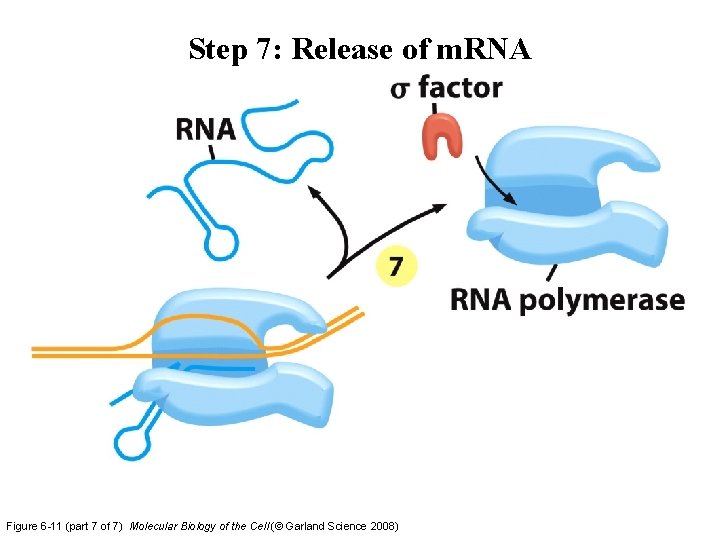
Step 7: Release of m. RNA Figure 6 -11 (part 7 of 7) Molecular Biology of the Cell (© Garland Science 2008)
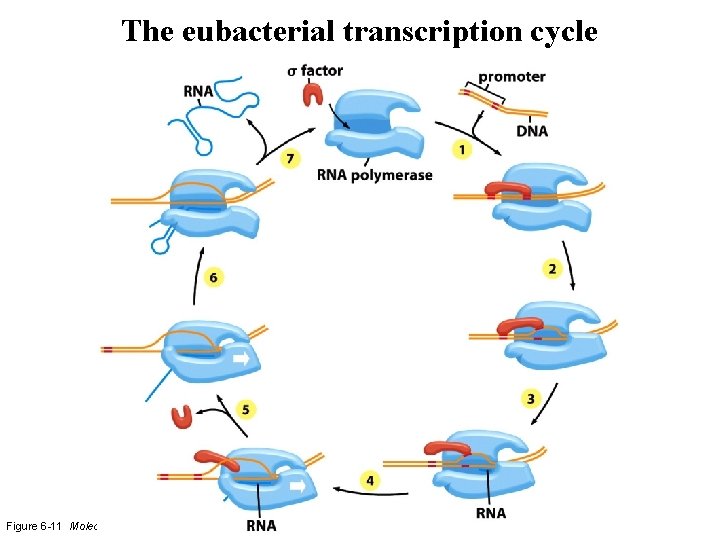
The eubacterial transcription cycle Figure 6 -11 Molecular Biology of the Cell (© Garland Science 2008)
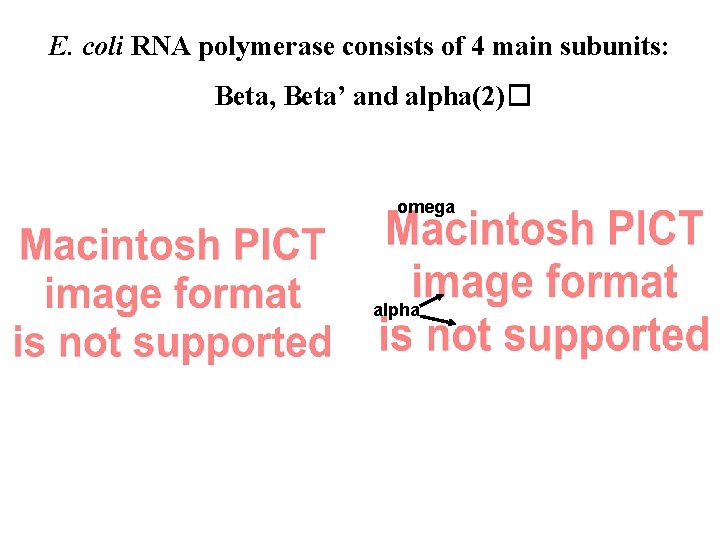
E. coli RNA polymerase consists of 4 main subunits: Beta, Beta’ and alpha(2)� omega alpha
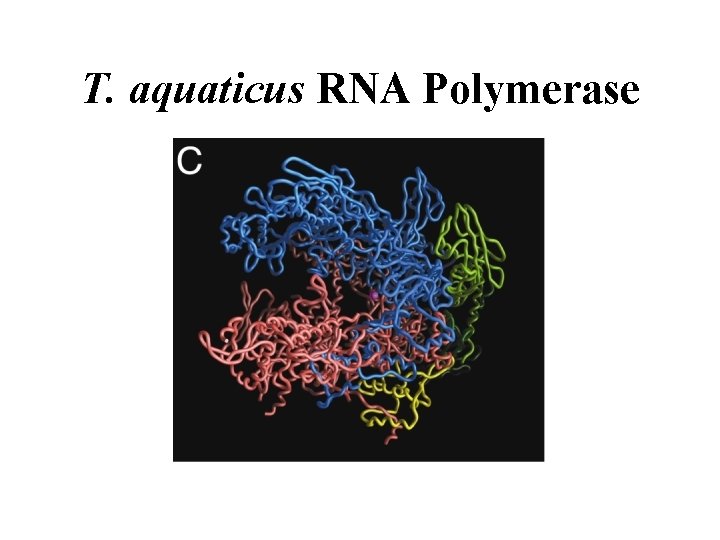
T. aquaticus RNA Polymerase
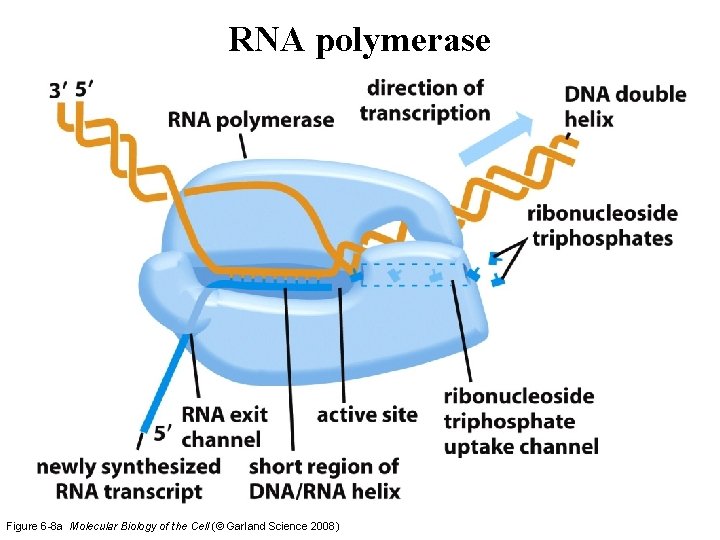
RNA polymerase Figure 6 -8 a Molecular Biology of the Cell (© Garland Science 2008)
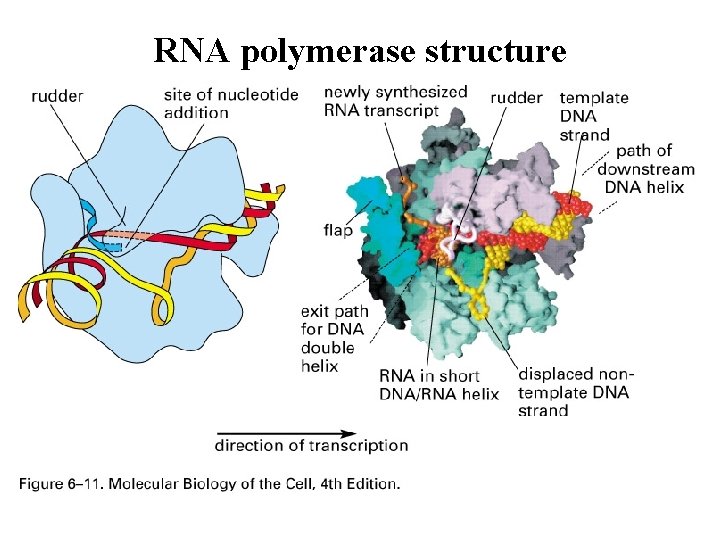
RNA polymerase structure
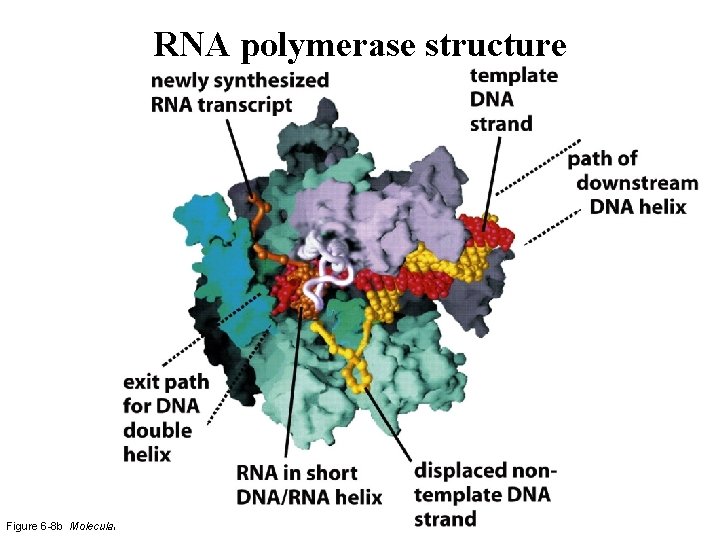
RNA polymerase structure Figure 6 -8 b Molecular Biology of the Cell (© Garland Science 2008)

RNA polymerase structure
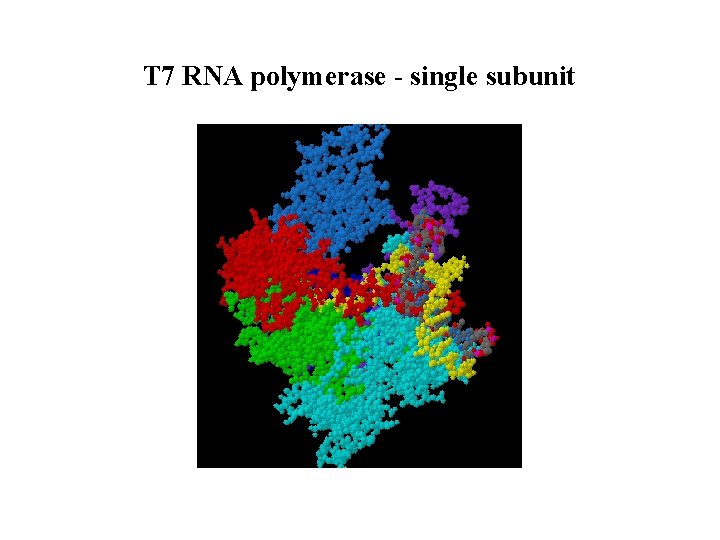
T 7 RNA polymerase - single subunit
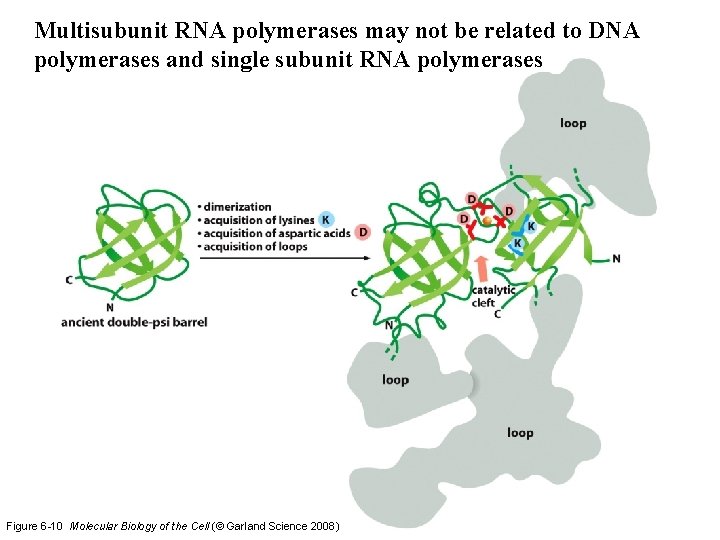
Multisubunit RNA polymerases may not be related to DNA polymerases and single subunit RNA polymerases Figure 6 -10 Molecular Biology of the Cell (© Garland Science 2008)
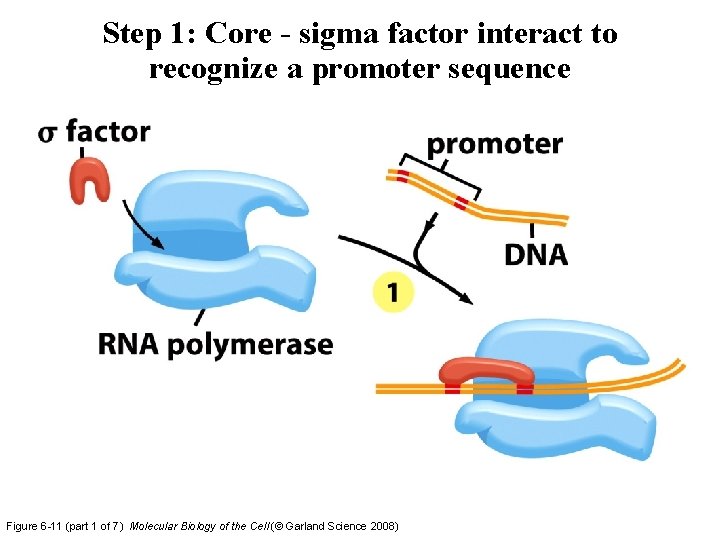
Step 1: Core - sigma factor interact to recognize a promoter sequence Figure 6 -11 (part 1 of 7) Molecular Biology of the Cell (© Garland Science 2008)
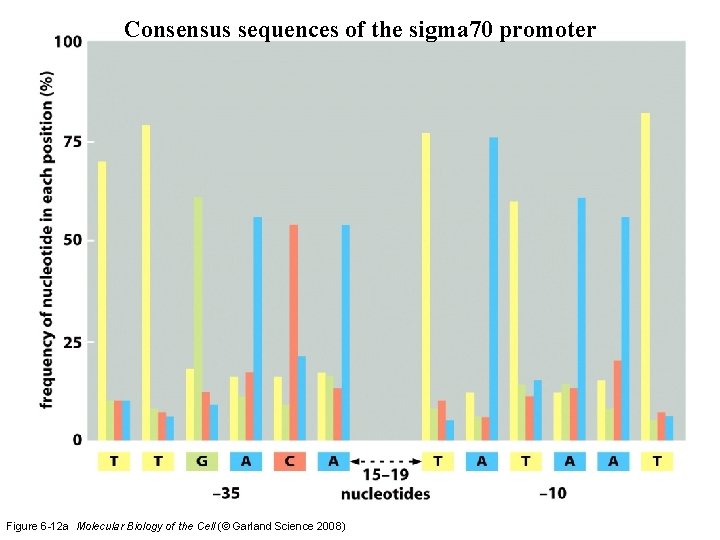
Consensus sequences of the sigma 70 promoter Figure 6 -12 a Molecular Biology of the Cell (© Garland Science 2008)
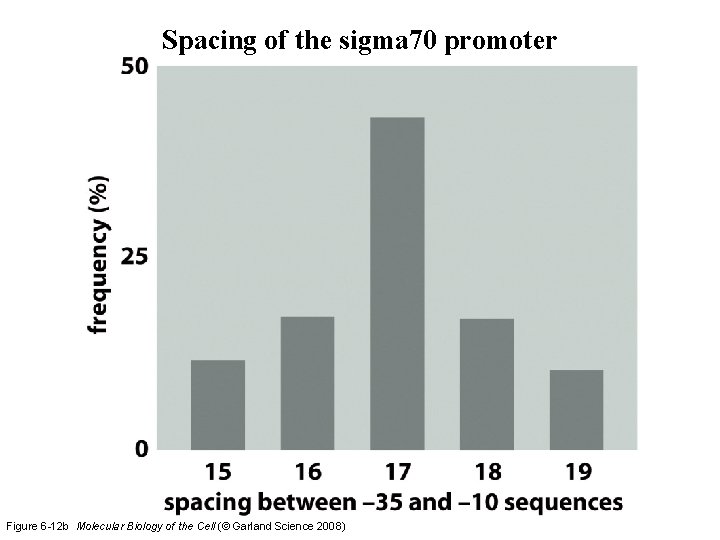
Spacing of the sigma 70 promoter Figure 6 -12 b Molecular Biology of the Cell (© Garland Science 2008)
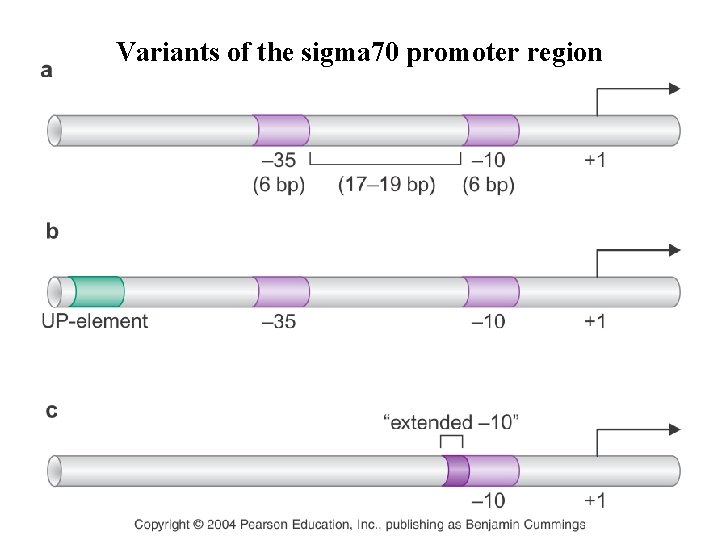
Variants of the sigma 70 promoter region
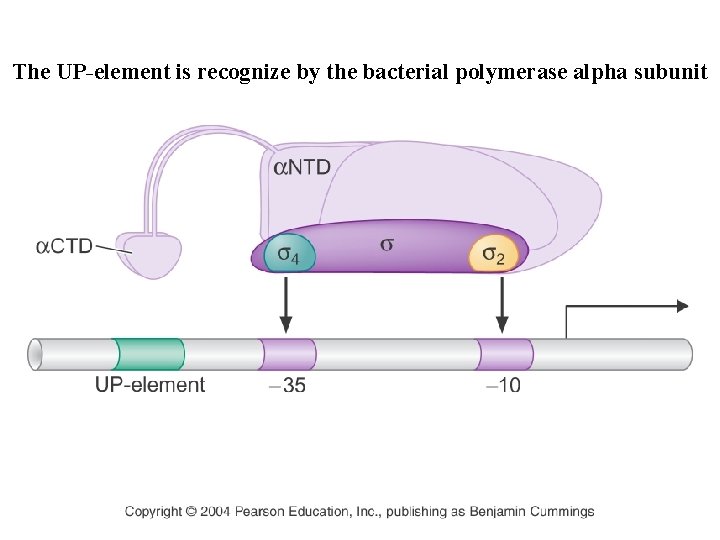
The UP-element is recognize by the bacterial polymerase alpha subunit
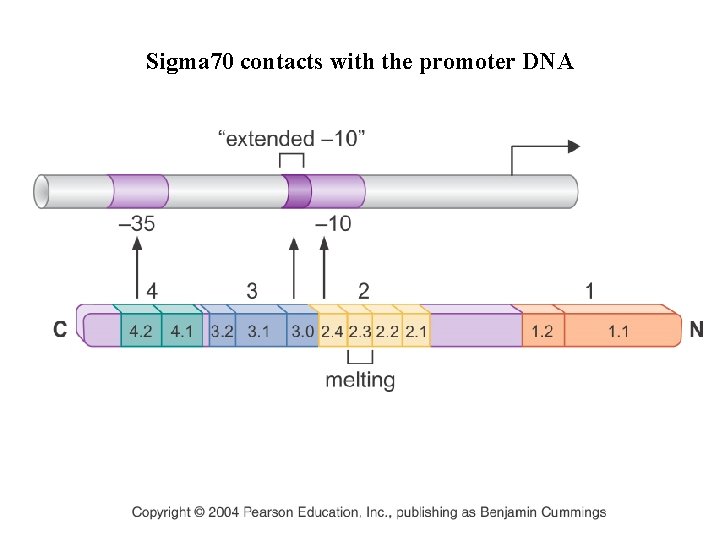
Sigma 70 contacts with the promoter DNA
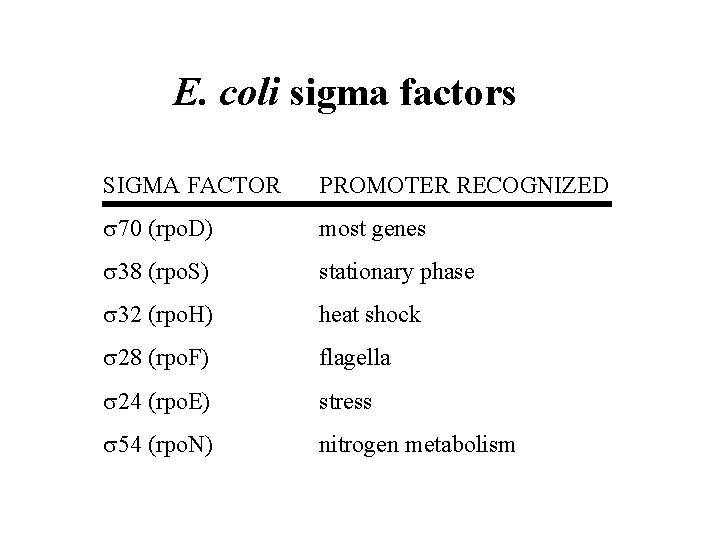
E. coli sigma factors SIGMA FACTOR PROMOTER RECOGNIZED 70 (rpo. D) most genes 38 (rpo. S) stationary phase 32 (rpo. H) heat shock 28 (rpo. F) flagella 24 (rpo. E) stress 54 (rpo. N) nitrogen metabolism
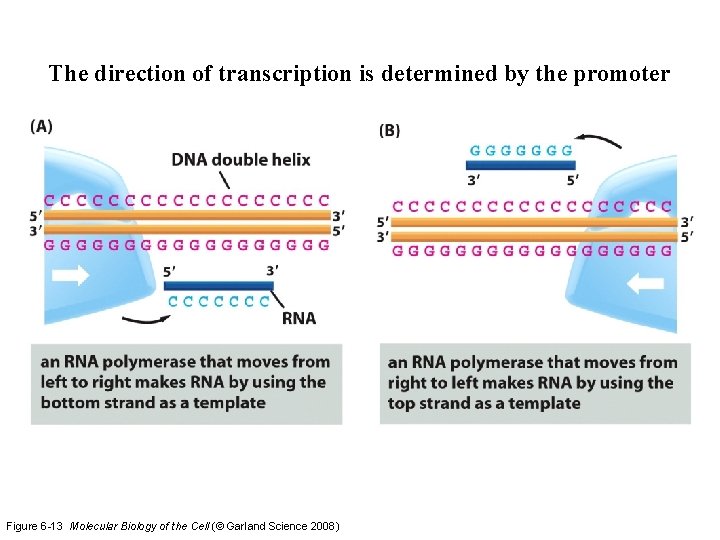
The direction of transcription is determined by the promoter Figure 6 -13 Molecular Biology of the Cell (© Garland Science 2008)
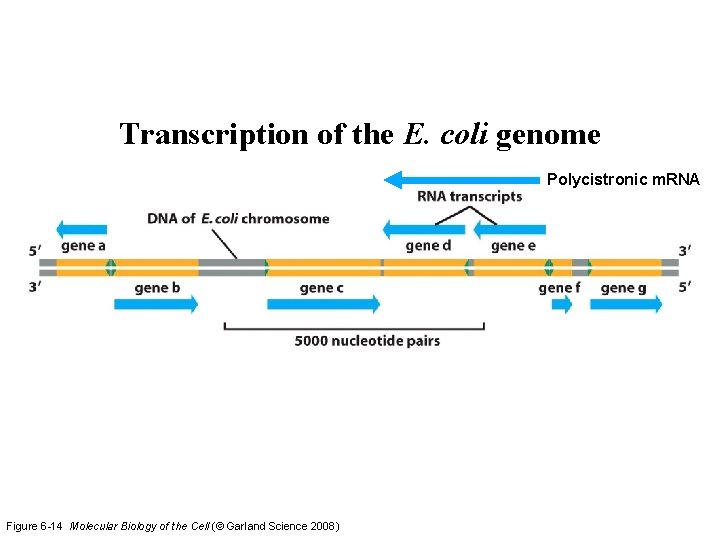
Transcription of the E. coli genome Polycistronic m. RNA Figure 6 -14 Molecular Biology of the Cell (© Garland Science 2008)
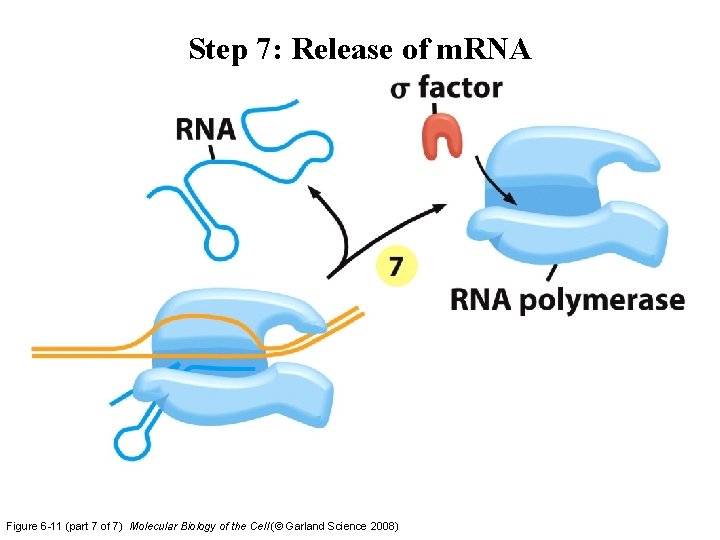
Step 7: Release of m. RNA Figure 6 -11 (part 7 of 7) Molecular Biology of the Cell (© Garland Science 2008)
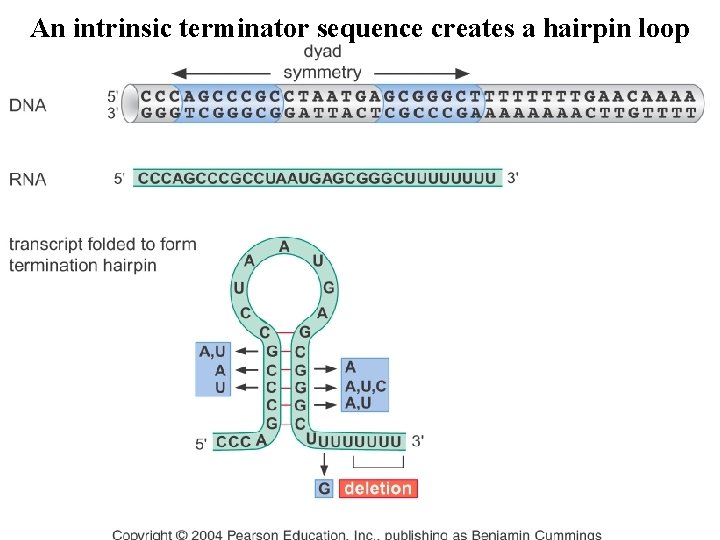
An intrinsic terminator sequence creates a hairpin loop
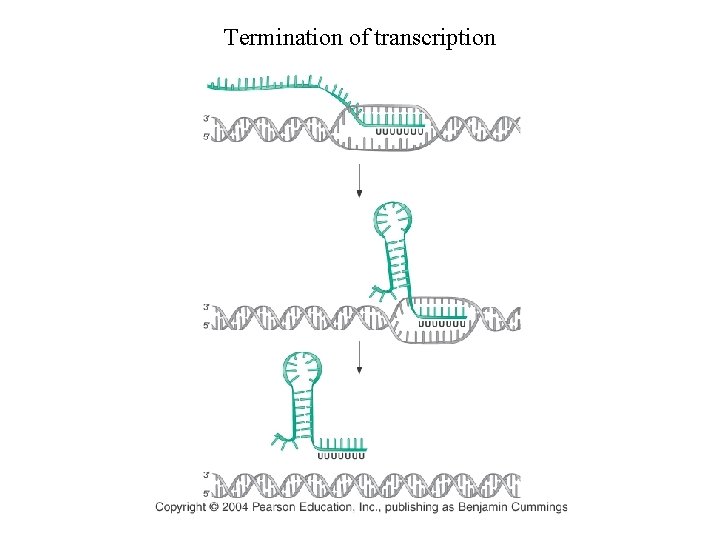
Termination of transcription
Rho terminator
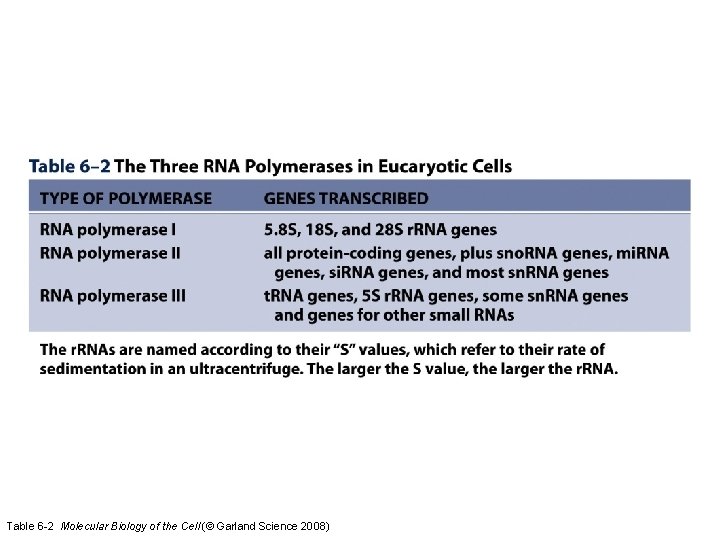
Table 6 -2 Molecular Biology of the Cell (© Garland Science 2008)
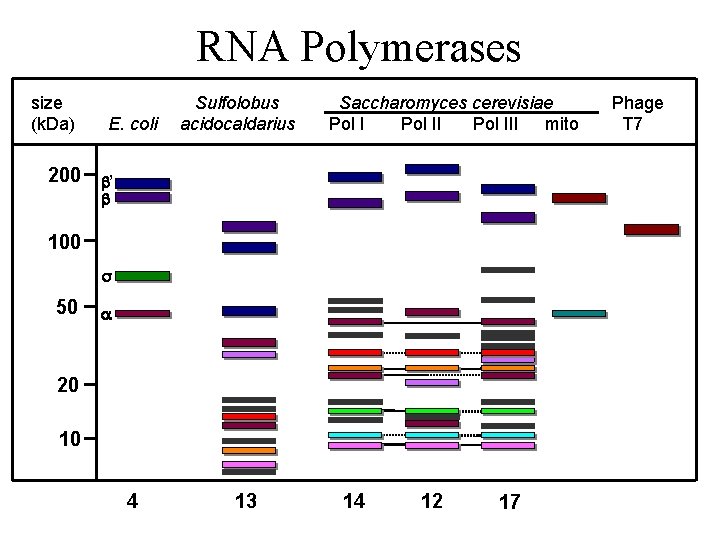
RNA Polymerases size (k. Da) 200 E. coli Sulfolobus acidocaldarius Saccharomyces cerevisiae Pol III mito ’ 100 50 20 10 4 13 14 12 17 Phage T 7
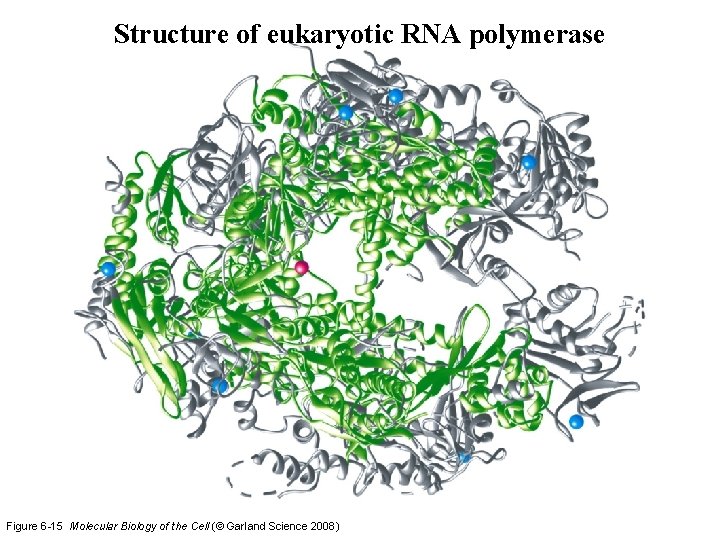
Structure of eukaryotic RNA polymerase Figure 6 -15 Molecular Biology of the Cell (© Garland Science 2008)
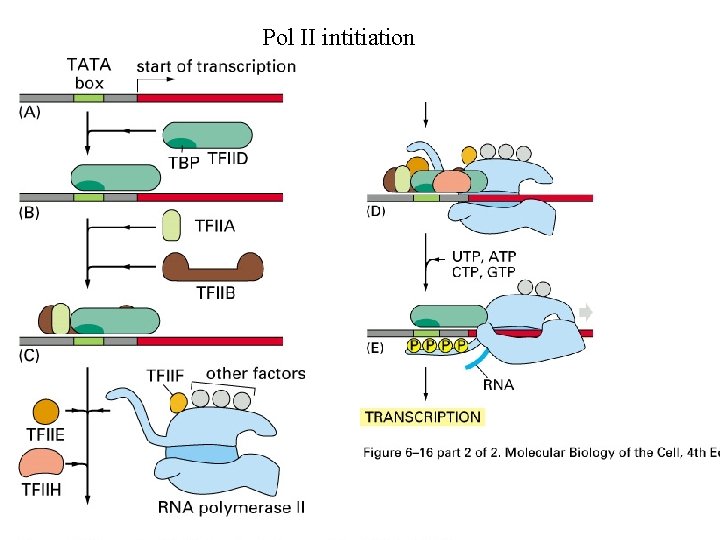
Pol II intitiation
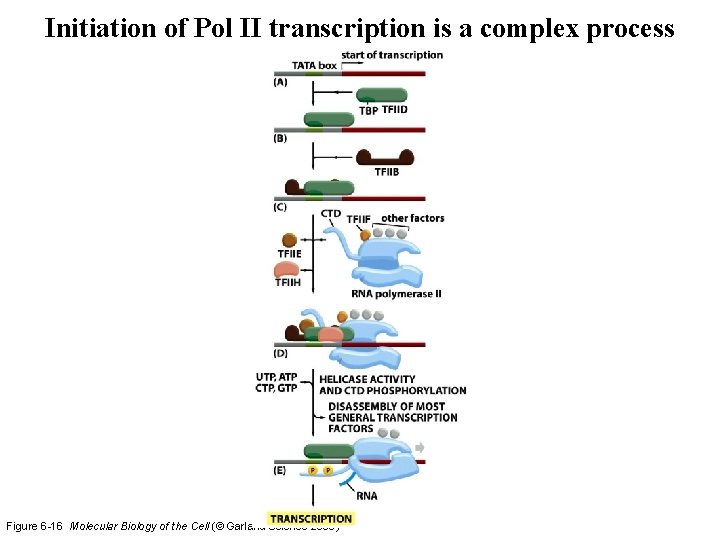
Initiation of Pol II transcription is a complex process Figure 6 -16 Molecular Biology of the Cell (© Garland Science 2008)
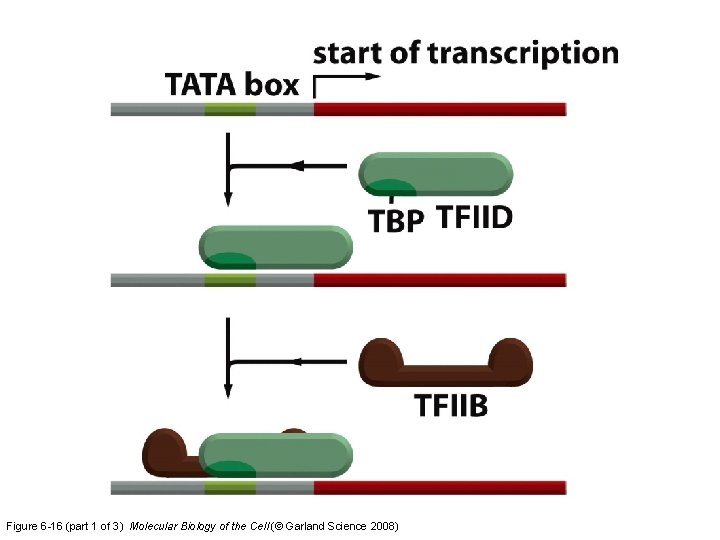
Figure 6 -16 (part 1 of 3) Molecular Biology of the Cell (© Garland Science 2008)
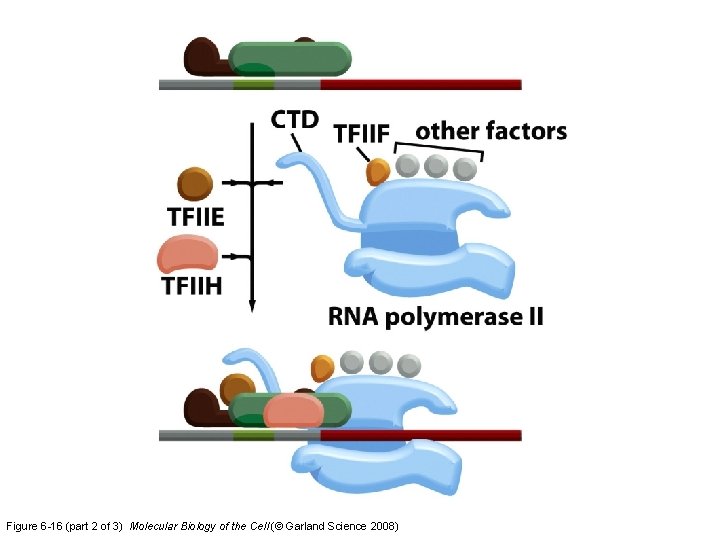
Figure 6 -16 (part 2 of 3) Molecular Biology of the Cell (© Garland Science 2008)
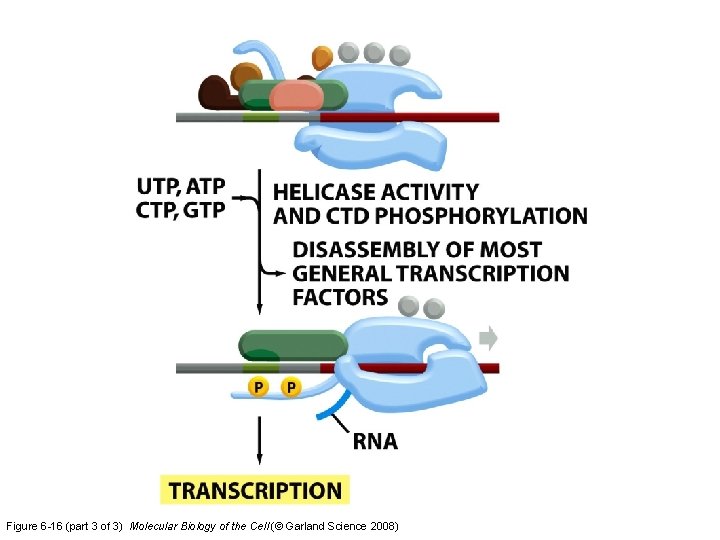
Figure 6 -16 (part 3 of 3) Molecular Biology of the Cell (© Garland Science 2008)
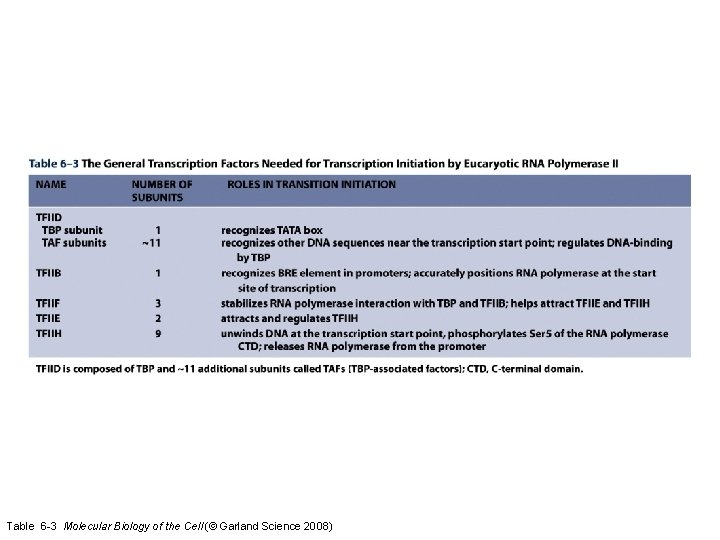
Table 6 -3 Molecular Biology of the Cell (© Garland Science 2008)
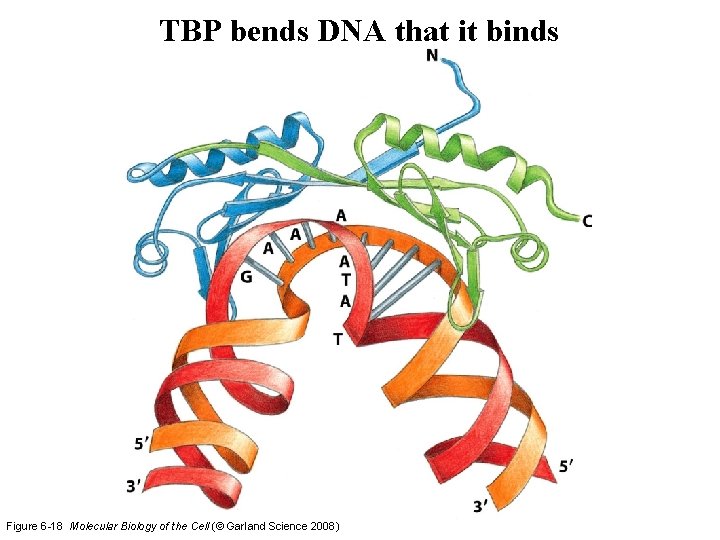
TBP bends DNA that it binds Figure 6 -18 Molecular Biology of the Cell (© Garland Science 2008)
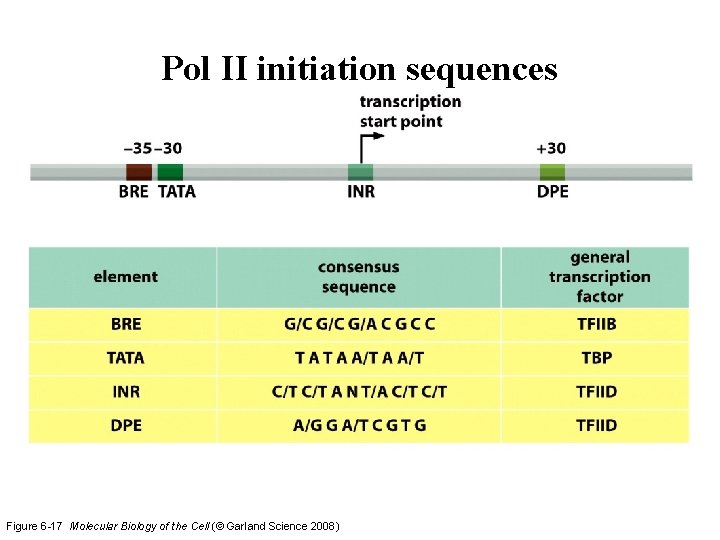
Pol II initiation sequences Figure 6 -17 Molecular Biology of the Cell (© Garland Science 2008)
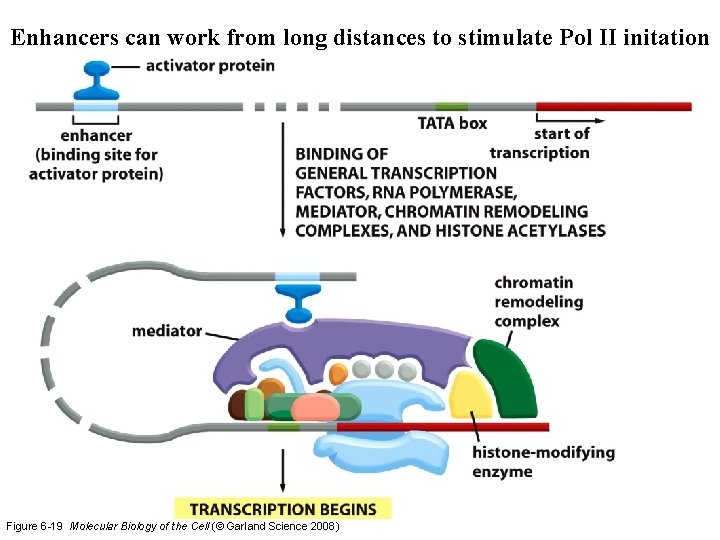
Enhancers can work from long distances to stimulate Pol II initation Figure 6 -19 Molecular Biology of the Cell (© Garland Science 2008)
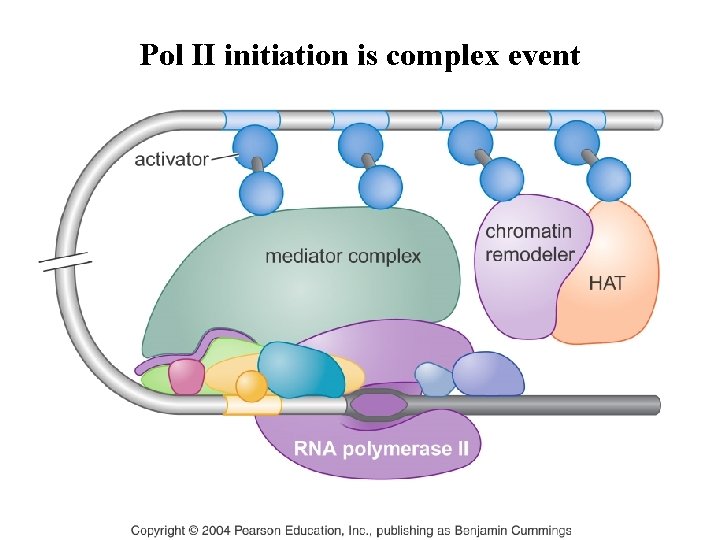
Pol II initiation is complex event
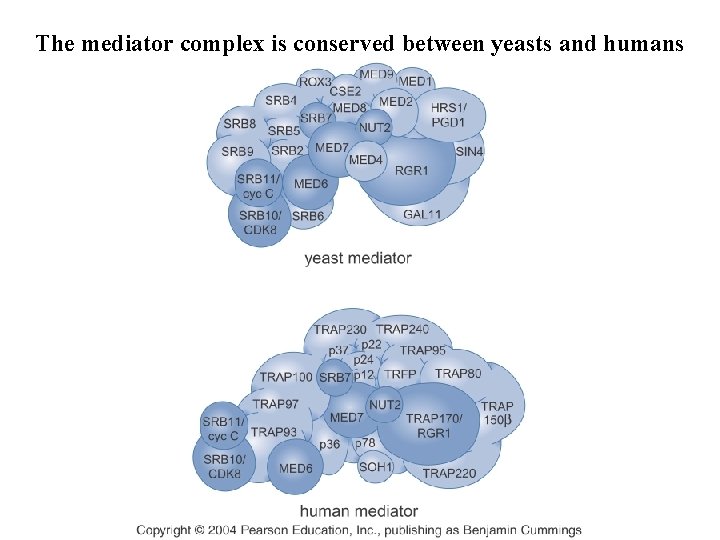
The mediator complex is conserved between yeasts and humans
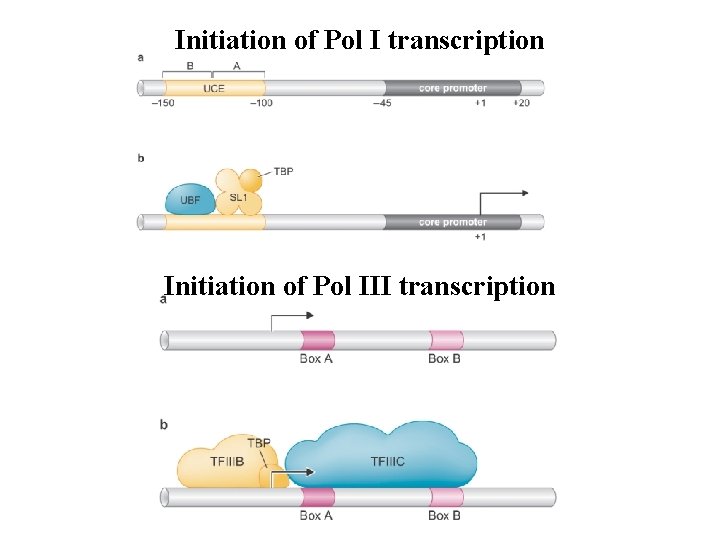
Initiation of Pol I transcription Initiation of Pol III transcription

Elongation
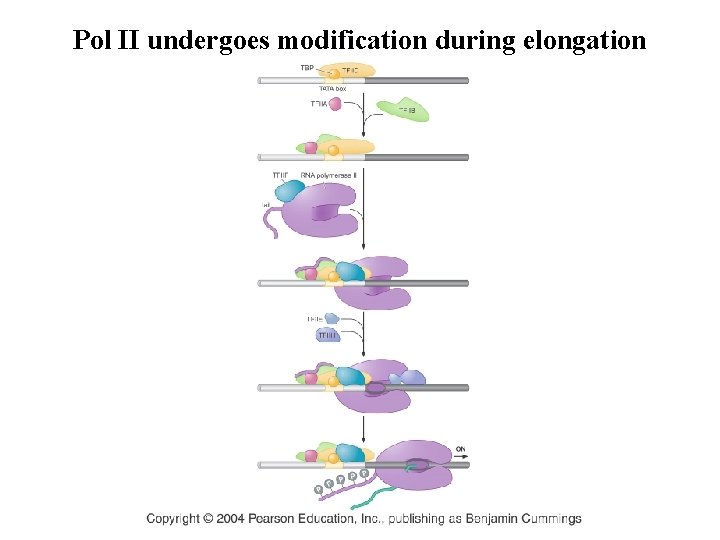
Pol II undergoes modification during elongation
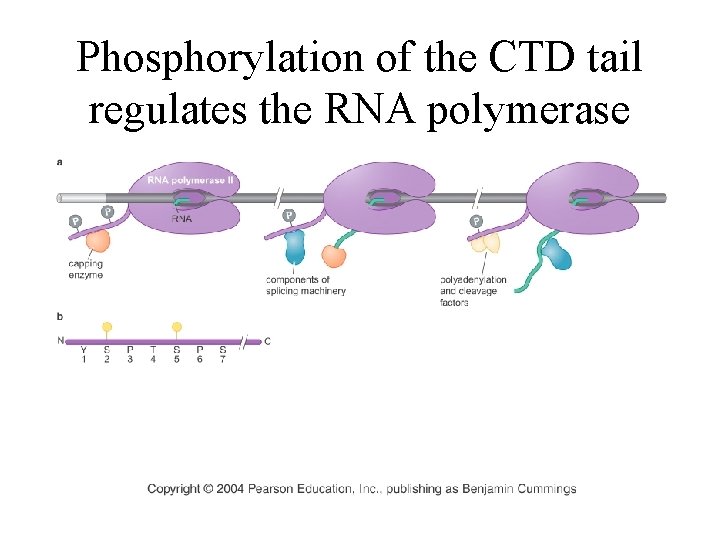
Phosphorylation of the CTD tail regulates the RNA polymerase
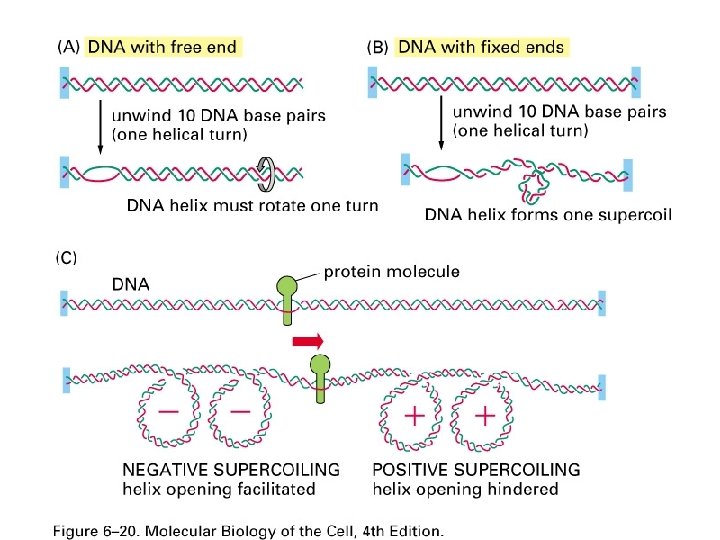
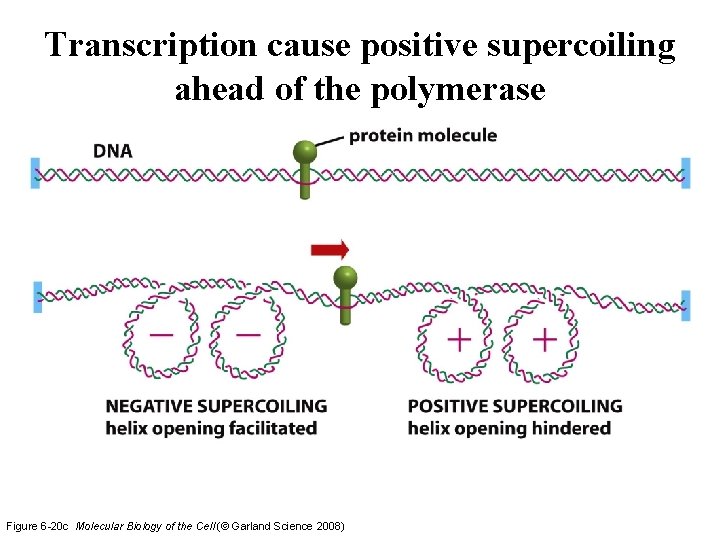
Transcription cause positive supercoiling ahead of the polymerase Figure 6 -20 c Molecular Biology of the Cell (© Garland Science 2008)

Post-transcriptional modification of m. RNA
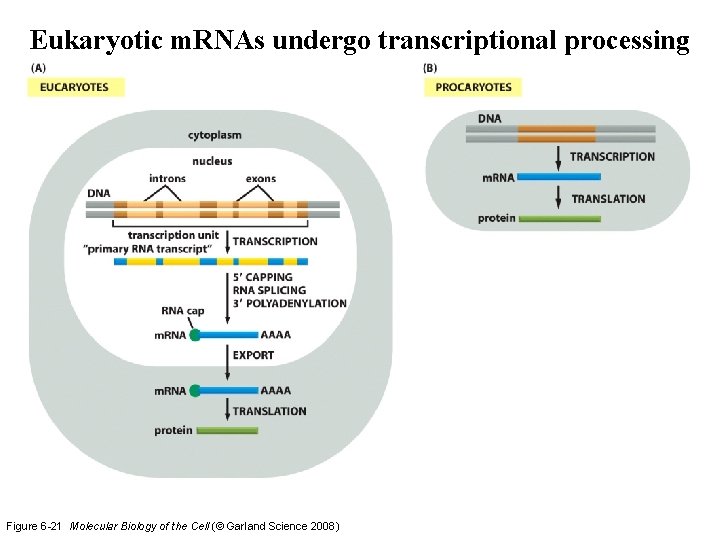
Eukaryotic m. RNAs undergo transcriptional processing Figure 6 -21 Molecular Biology of the Cell (© Garland Science 2008)
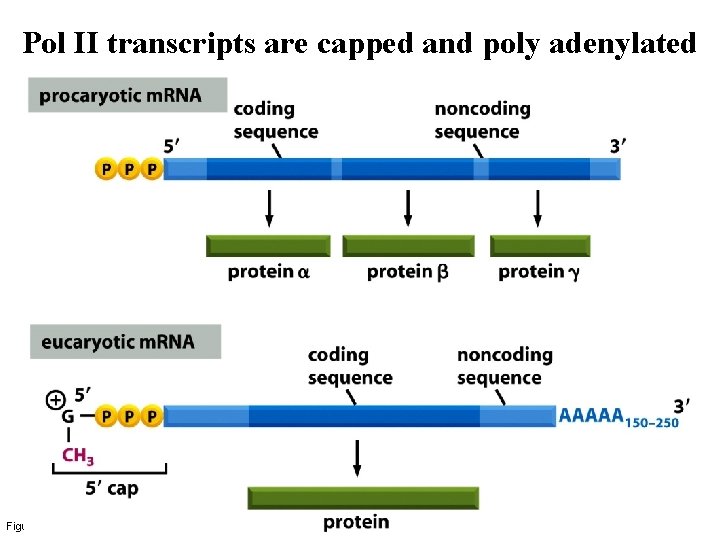
Pol II transcripts are capped and poly adenylated Figure 6 -22 a Molecular Biology of the Cell (© Garland Science 2008)
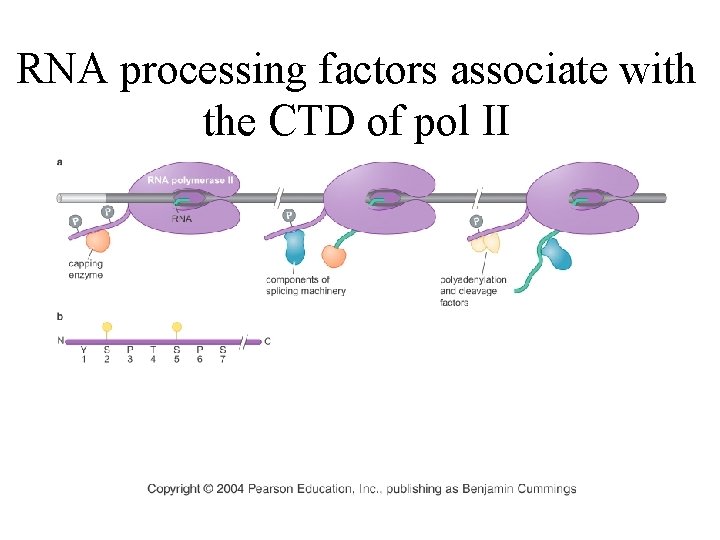
RNA processing factors associate with the CTD of pol II
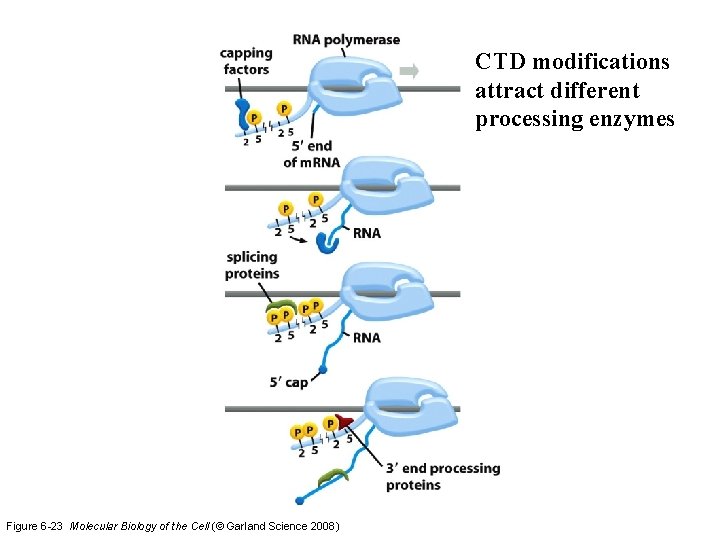
CTD modifications attract different processing enzymes Figure 6 -23 Molecular Biology of the Cell (© Garland Science 2008)
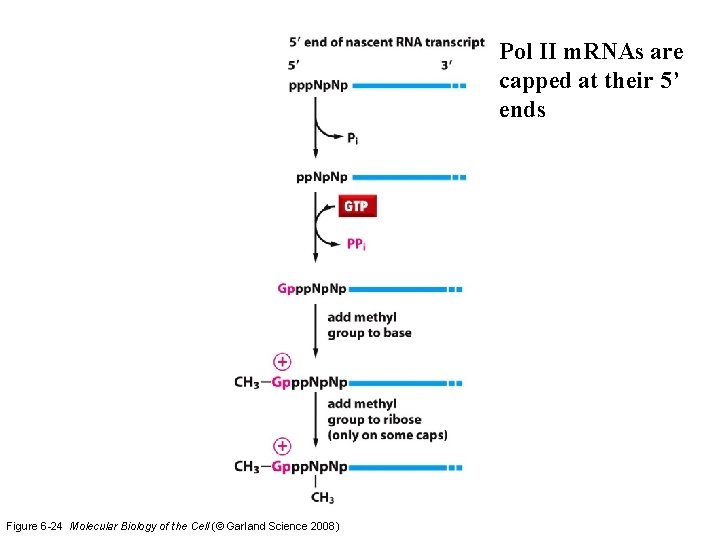
Pol II m. RNAs are capped at their 5’ ends Figure 6 -24 Molecular Biology of the Cell (© Garland Science 2008)
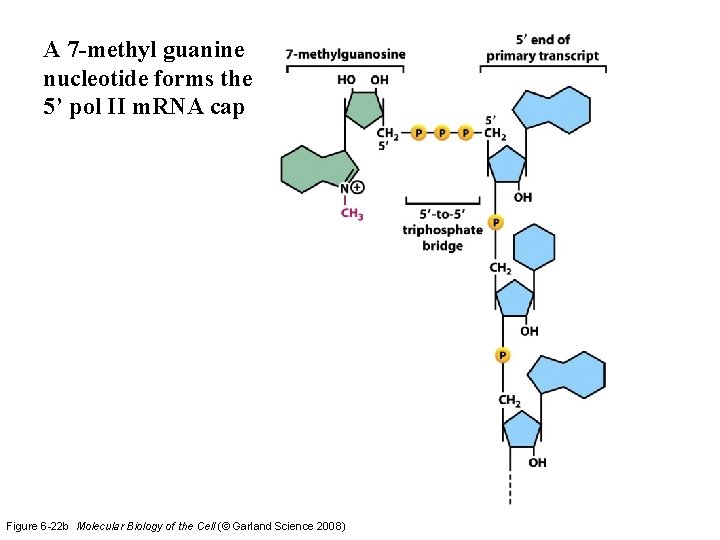
A 7 -methyl guanine nucleotide forms the 5’ pol II m. RNA cap Figure 6 -22 b Molecular Biology of the Cell (© Garland Science 2008)
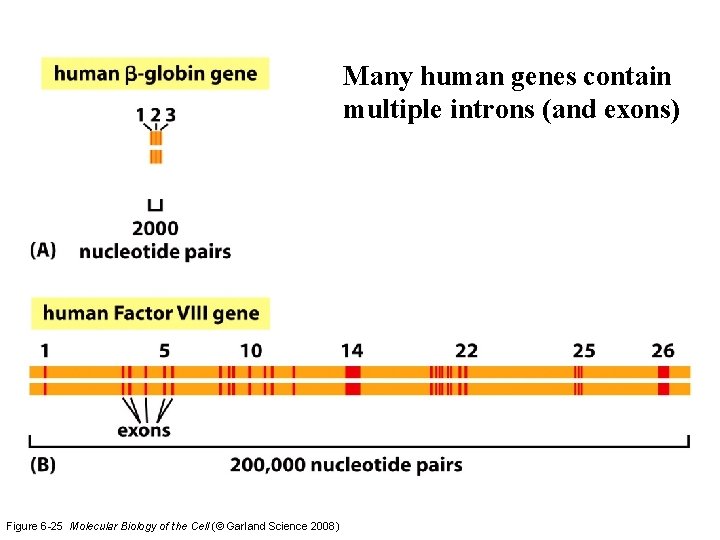
Many human genes contain multiple introns (and exons) Figure 6 -25 Molecular Biology of the Cell (© Garland Science 2008)
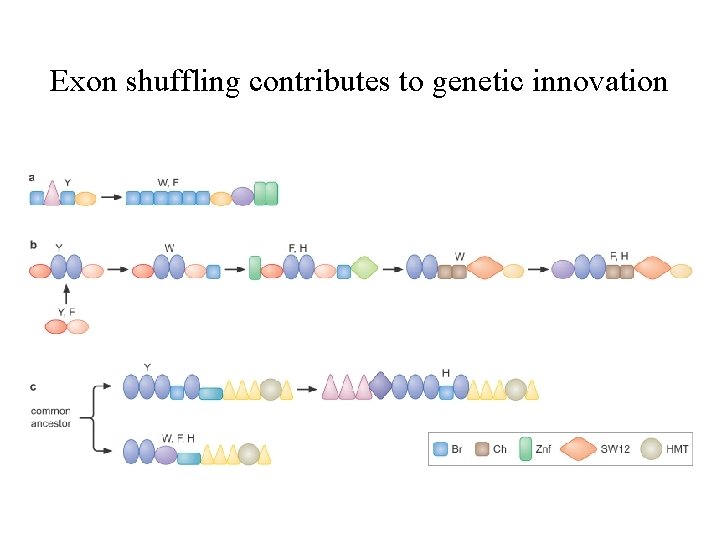
Exon shuffling contributes to genetic innovation
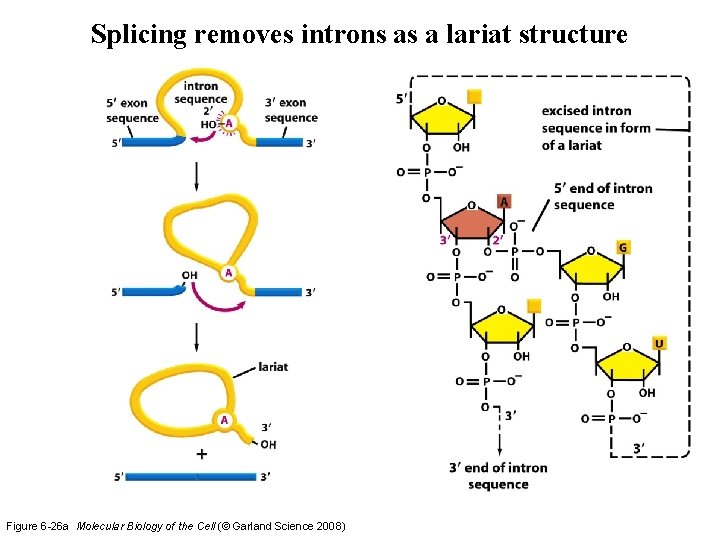
Splicing removes introns as a lariat structure Figure 6 -26 a Molecular Biology of the Cell (© Garland Science 2008)
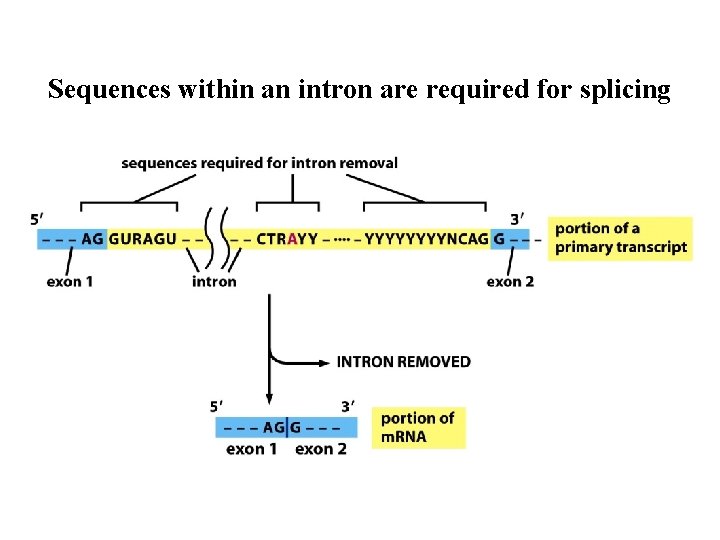
Sequences within an intron are required for splicing
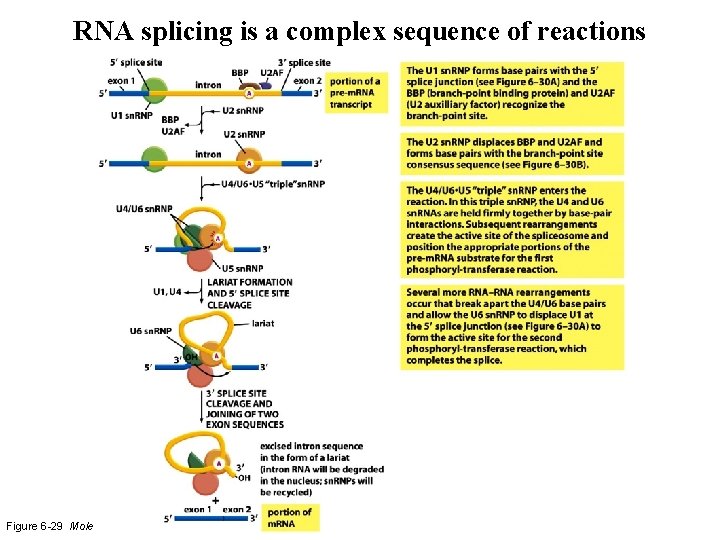
RNA splicing is a complex sequence of reactions Figure 6 -29 Molecular Biology of the Cell (© Garland Science 2008)
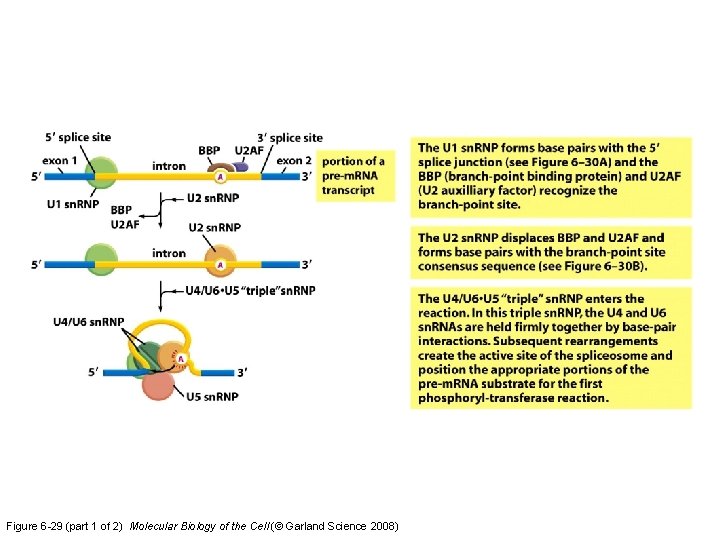
Figure 6 -29 (part 1 of 2) Molecular Biology of the Cell (© Garland Science 2008)
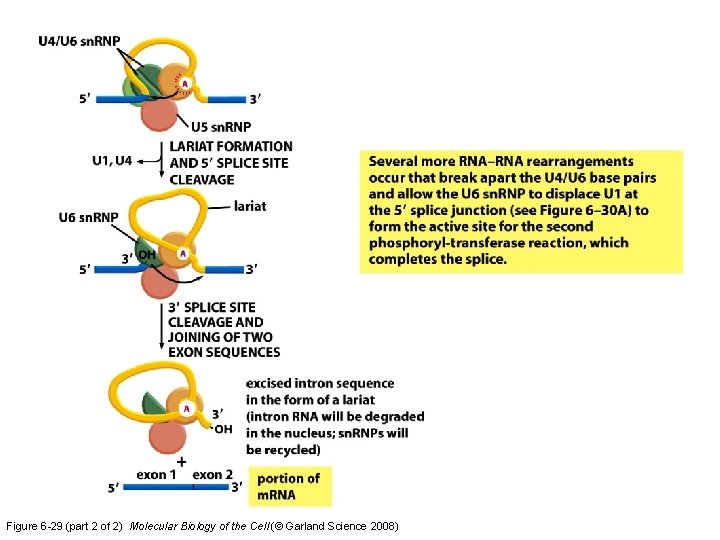
Figure 6 -29 (part 2 of 2) Molecular Biology of the Cell (© Garland Science 2008)
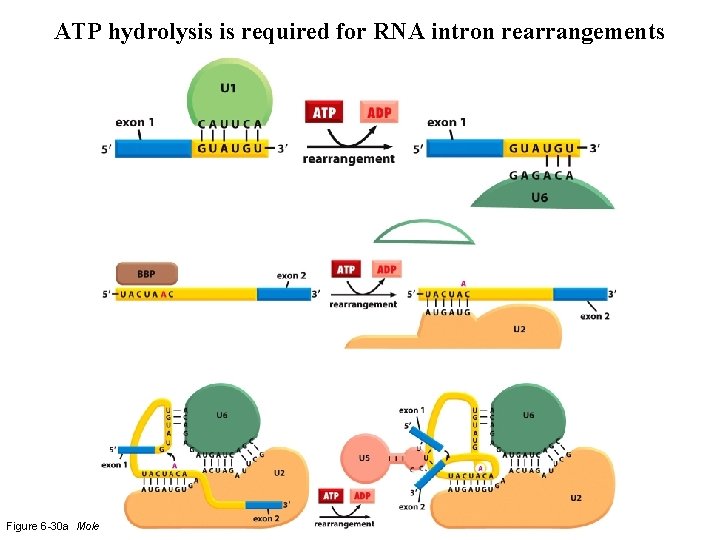
ATP hydrolysis is required for RNA intron rearrangements Figure 6 -30 a Molecular Biology of the Cell (© Garland Science 2008)
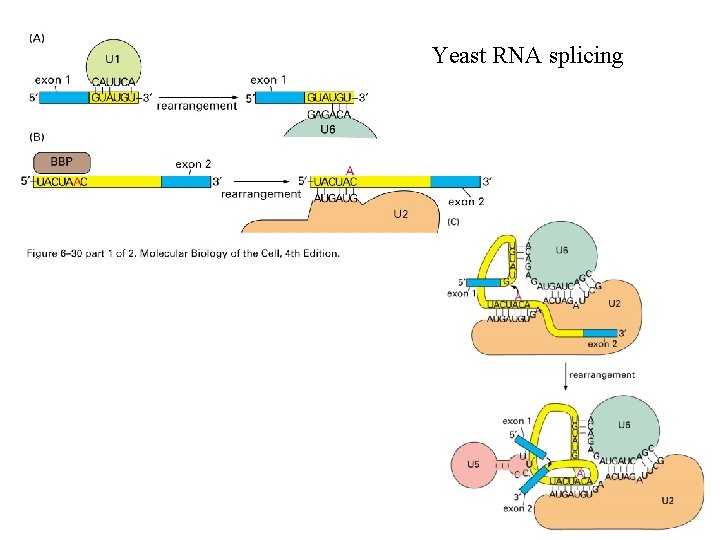
Yeast RNA splicing
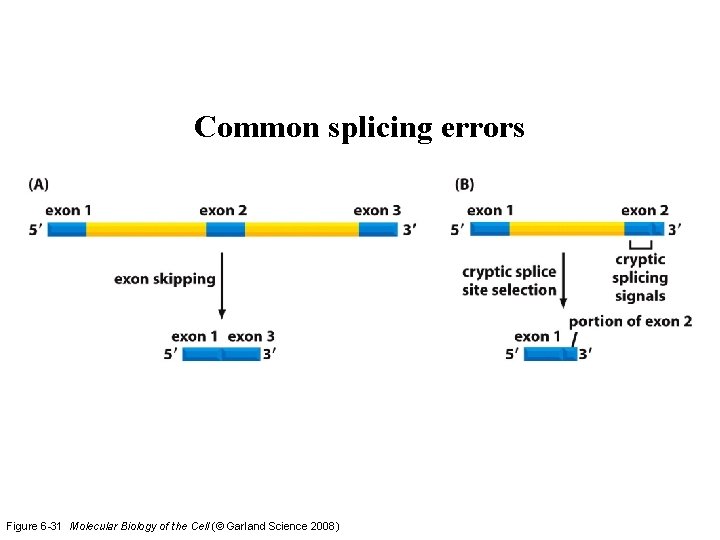
Common splicing errors Figure 6 -31 Molecular Biology of the Cell (© Garland Science 2008)
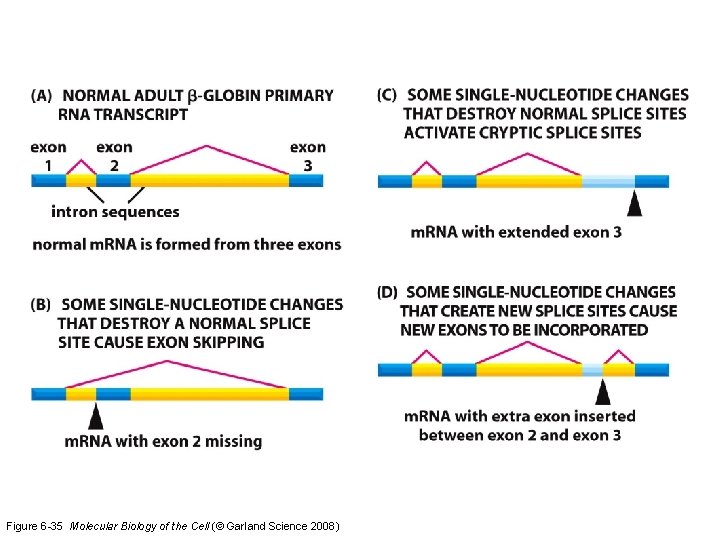
Figure 6 -35 Molecular Biology of the Cell (© Garland Science 2008)

The exon definition hypothesis is a model for how exons are recognized in a m. RNA Figure 6 -33 Molecular Biology of the Cell (© Garland Science 2008)
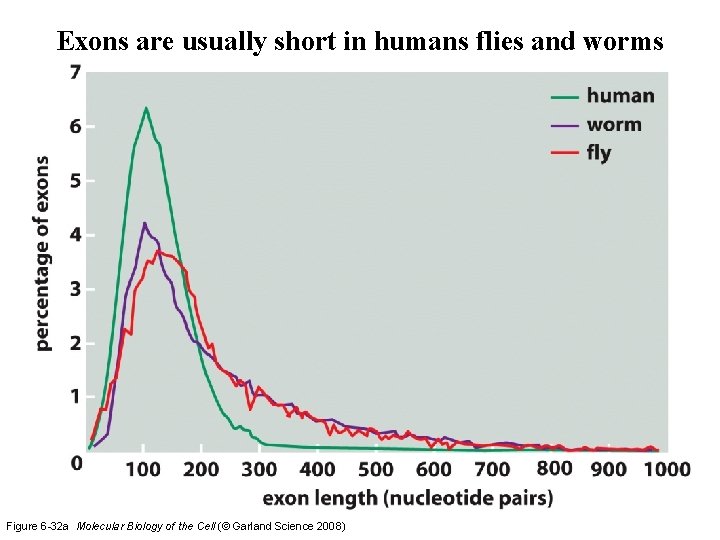
Exons are usually short in humans flies and worms Figure 6 -32 a Molecular Biology of the Cell (© Garland Science 2008)
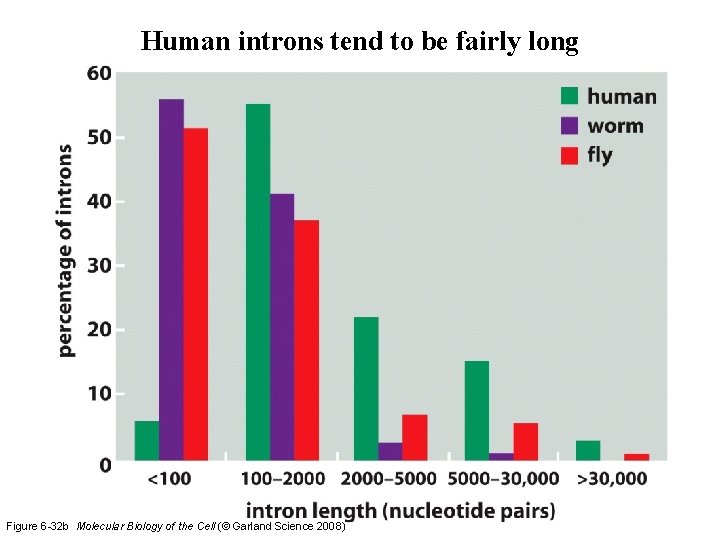
Human introns tend to be fairly long Figure 6 -32 b Molecular Biology of the Cell (© Garland Science 2008)
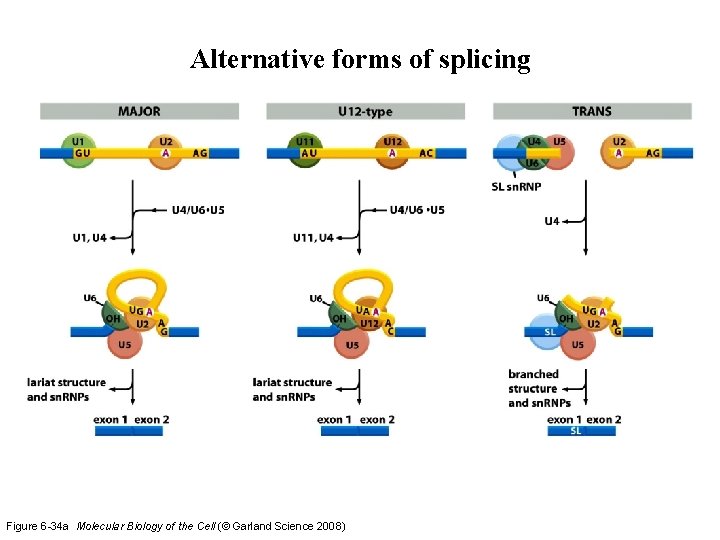
Alternative forms of splicing Figure 6 -34 a Molecular Biology of the Cell (© Garland Science 2008)
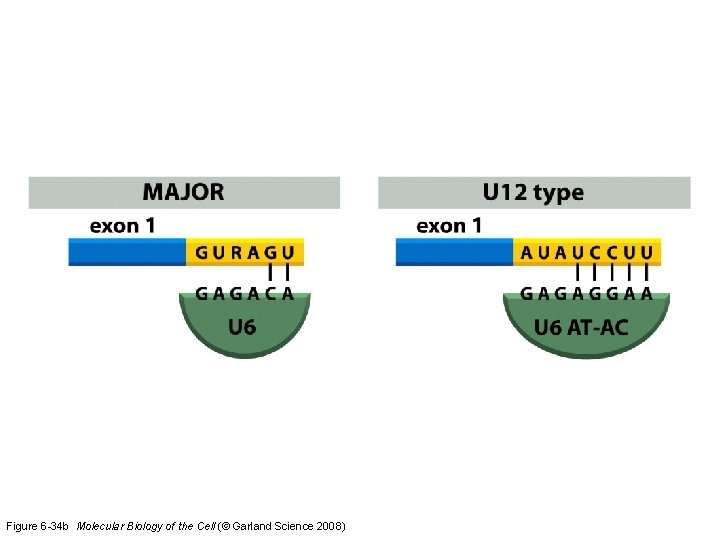
Figure 6 -34 b Molecular Biology of the Cell (© Garland Science 2008)
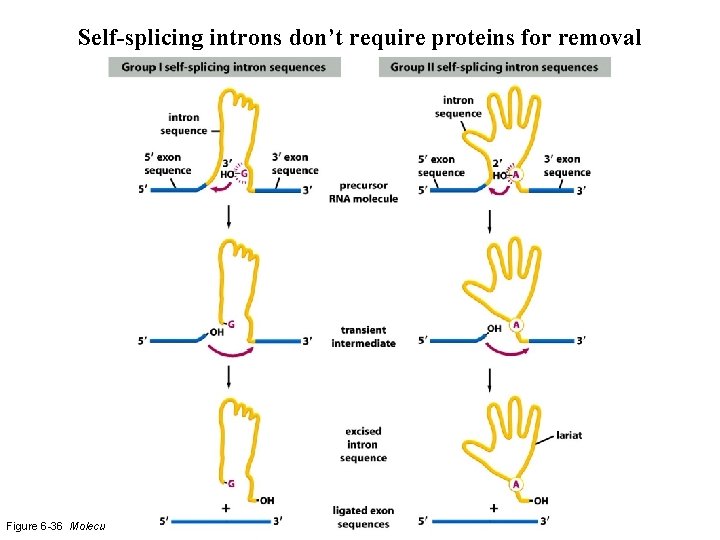
Self-splicing introns don’t require proteins for removal Figure 6 -36 Molecular Biology of the Cell (© Garland Science 2008)
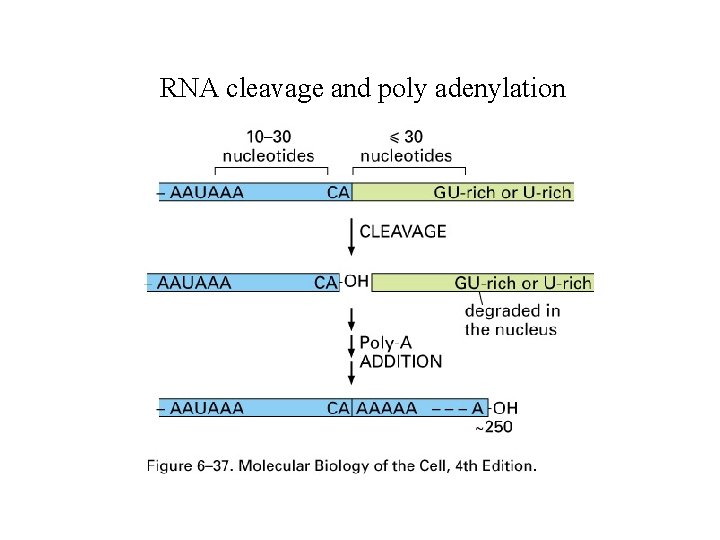
RNA cleavage and poly adenylation
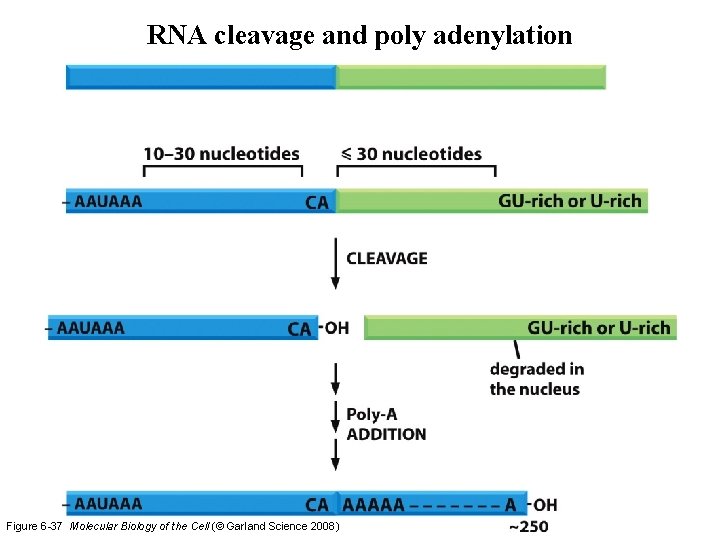
RNA cleavage and poly adenylation Figure 6 -37 Molecular Biology of the Cell (© Garland Science 2008)
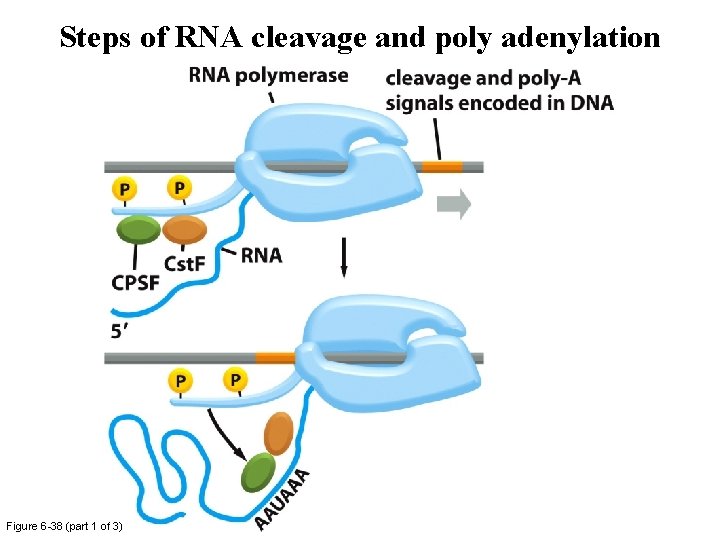
Steps of RNA cleavage and poly adenylation Figure 6 -38 (part 1 of 3)
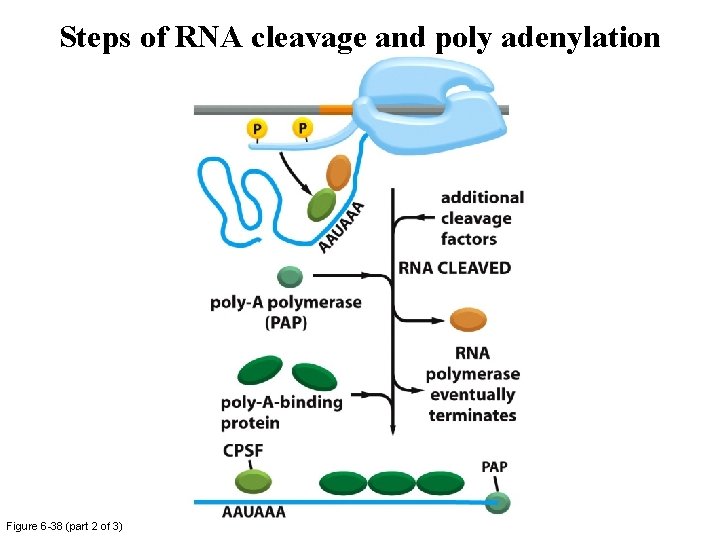
Steps of RNA cleavage and poly adenylation Figure 6 -38 (part 2 of 3)
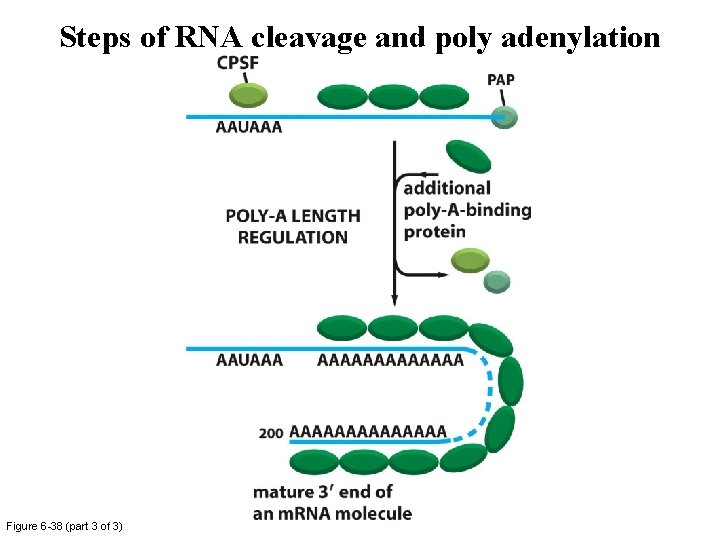
Steps of RNA cleavage and poly adenylation Figure 6 -38 (part 3 of 3)
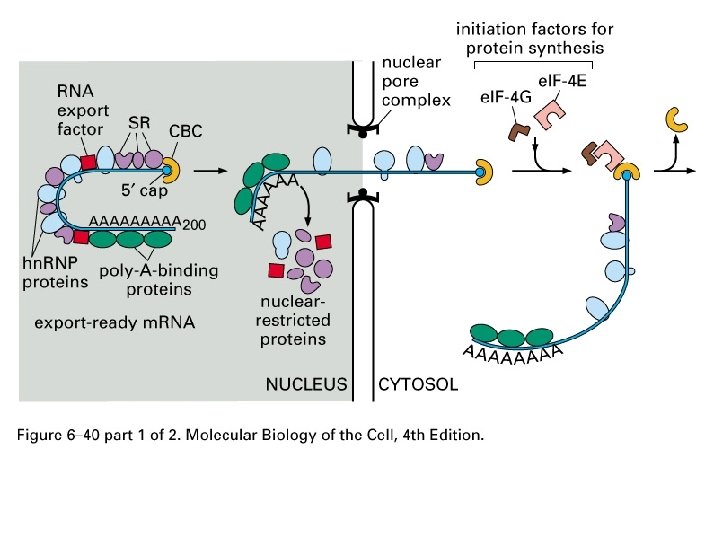
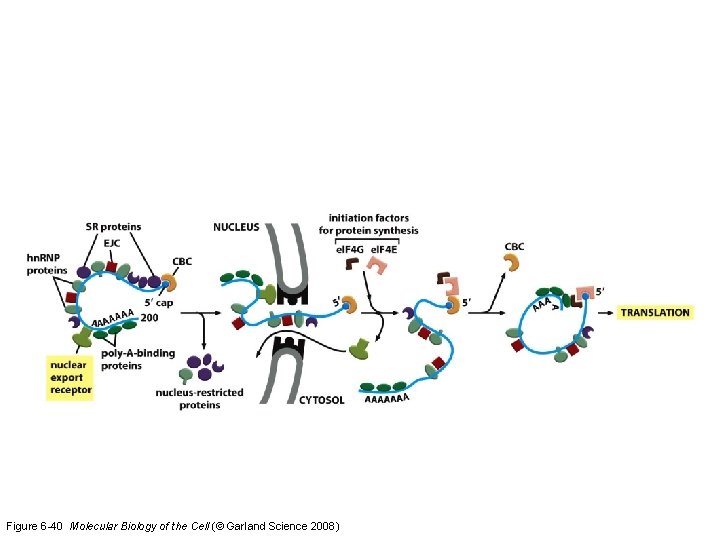
Figure 6 -40 Molecular Biology of the Cell (© Garland Science 2008)
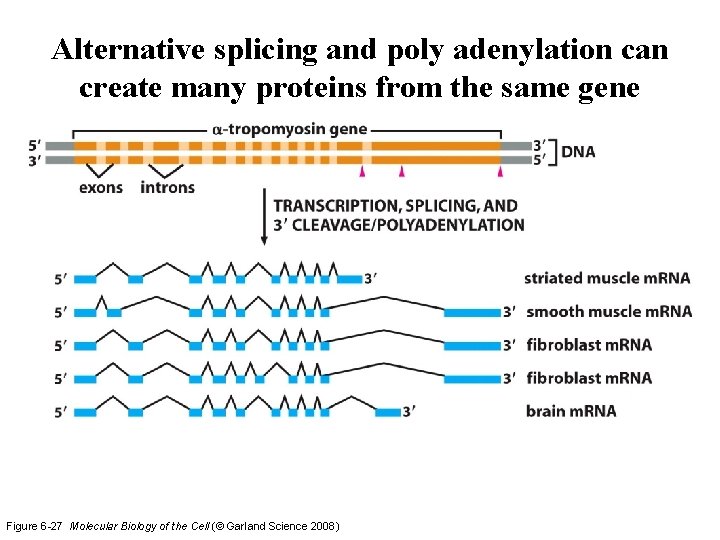
Alternative splicing and poly adenylation can create many proteins from the same gene Figure 6 -27 Molecular Biology of the Cell (© Garland Science 2008)

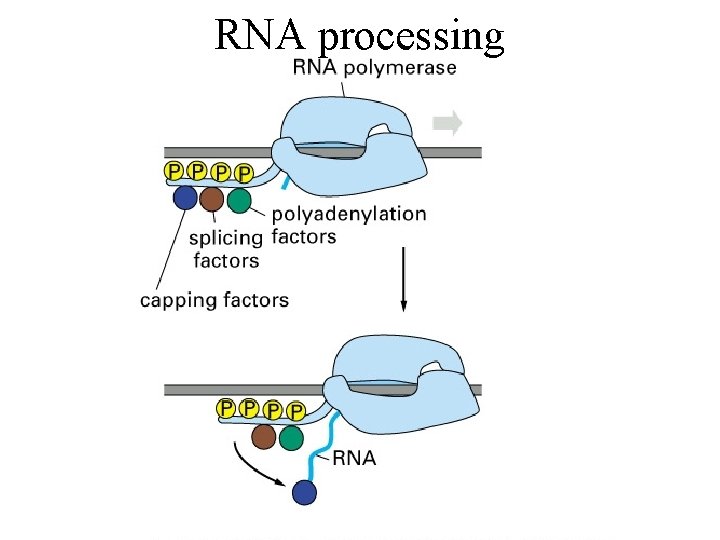
RNA processing
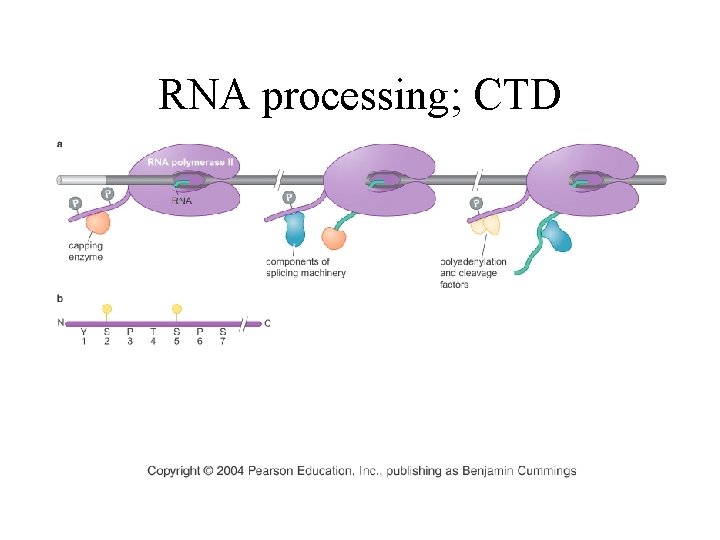
RNA processing; CTD
NOT COVERED m. RNA export Noncoding RNAS Nucleolus Subnuclear structures

Review Questions: What are the steps of spliceosomal removal of introns? What are two mechanisms of preventing splicing mistakes? What are the important features of the exon definition hypothesis? What are the differences between group I and group II intron splicing? What energy source is needed for spliceosomal splicing? What is it used for. What do sn. RNAs do? True or False There are over 50 spliceosomal proteins and each sn. RNA associates with at least 7 proteins?
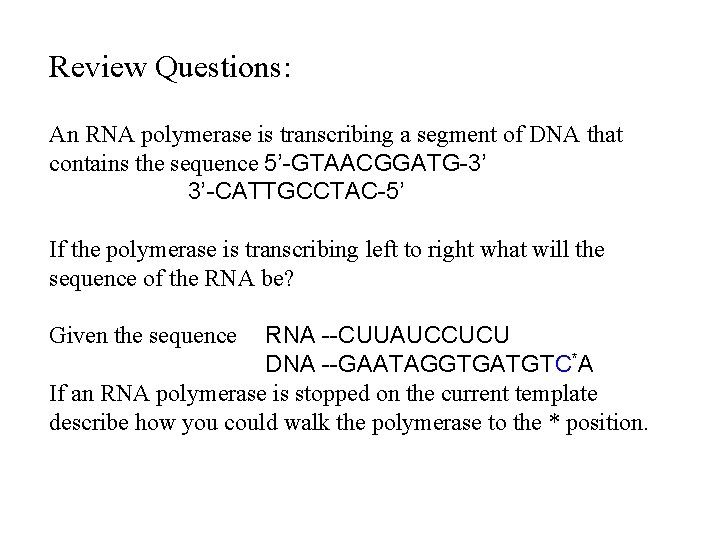
Review Questions: An RNA polymerase is transcribing a segment of DNA that contains the sequence 5’-GTAACGGATG-3’ 3’-CATTGCCTAC-5’ If the polymerase is transcribing left to right what will the sequence of the RNA be? RNA --CUUAUCCUCU DNA --GAATAGGTGATGTC*A If an RNA polymerase is stopped on the current template describe how you could walk the polymerase to the * position. Given the sequence
- Slides: 103