Affinity chromatography What is Affinity Chromatography A chromatographic



- Slides: 14

Affinity chromatography

What is Affinity Chromatography? • A chromatographic technique for the separation of the compound on the basis of a reversible interaction between a compound a specific ligand. • Ideal for purification of compound from complex mixtures several thousand folds in concentrated form. • Originally developed for purification of enzymes • Also purifies nucleotides, nucleic acids, immunoglobins, membrane receptors, whole cells and cell fragments. • High selectivity and high resolution

Principle • When a complex mixture containing specific compound is passed through a column containing immobilized ligand, the specific compound will bind to the ligand. Ligand is immobilized to an insoluble solid matrix. • A biological reversible interaction between a compound a specific ligand occurs as follows: Compound (M) + Ligand (L) ML complex • This interaction occurs due to – Electrostatic or hydrophobic interactions – Van der Waals' forces or – Hydrogen bonding.

Principle… • Compounds not specifically bound are washed away with the buffer. • Compounds specifically bound are recovered from ligand by reversing the interaction • There are two methods of elution Specific Non-specific

Types of elution Non-specific elution: Change of p. H or ionic strength or polarity. • p. H shift using dilute acetic acid or ammonium hydroxide causes a change in the state of ionization of groups in the ligand the compound. • A change in ionic strength causes elution due to a disruption of the ligand-compound interaction. Specific elution: Use of soluble competitive ligand.

Matrix/ Resins • Most common matrices Cross-linked dextrans, agarose, polymethylacrylate, polyacrylamide, polystyrene, cellulose, porous glass and silica. • Matrix particles are uniform, spherical and rigid. • An ideal matrix must have the following characteristics: – Should possess chemical groups to which the ligand may be covalently coupled. – Should be stable during binding of the compound and its subsequent elution – Should interact only weakly with the compound to minimize non-specific adsorption – Should exhibit good flow properties

Ligand • Ligand must specifically bind to one particular compound. • Ligand that display group selectivity selected as it will bind to closely related groups of compounds that posses similar chemical specificity. • Ligand must possess a chemical group that should not reversibly bind the ligand to the compound but should attach the ligand covalently to the matrix. • Most common chemical groups are –NH 2, -COOH, -SH and –OH.

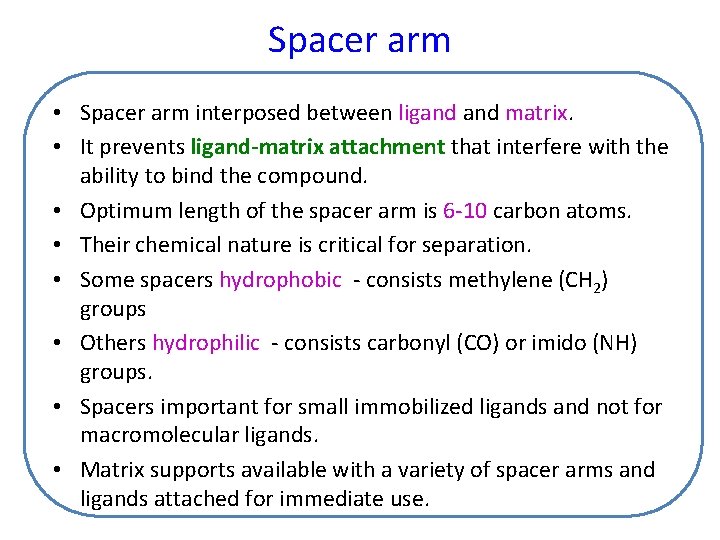
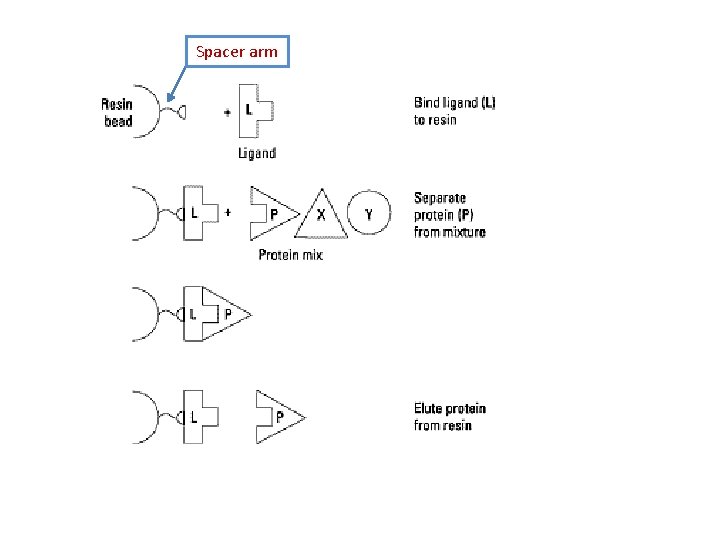
Spacer arm • Spacer arm interposed between ligand matrix. • It prevents ligand-matrix attachment that interfere with the ability to bind the compound. • Optimum length of the spacer arm is 6 -10 carbon atoms. • Their chemical nature is critical for separation. • Some spacers hydrophobic - consists methylene (CH 2) groups • Others hydrophilic - consists carbonyl (CO) or imido (NH) groups. • Spacers important for small immobilized ligands and not for macromolecular ligands. • Matrix supports available with a variety of spacer arms and ligands attached for immediate use.

Spacer arm

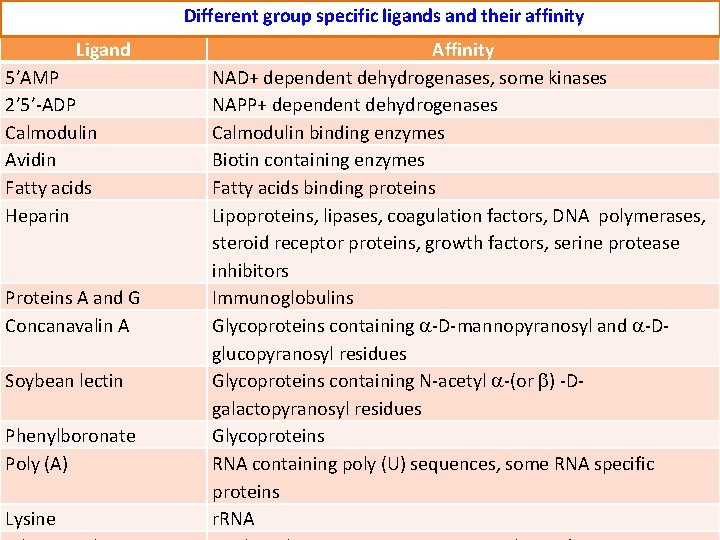
Different group specific ligands and their affinity. Ligand 5’AMP 2’ 5’-ADP Calmodulin Avidin Fatty acids Heparin Proteins A and G Concanavalin A Soybean lectin Phenylboronate Poly (A) Lysine Affinity NAD+ dependent dehydrogenases, some kinases NAPP+ dependent dehydrogenases Calmodulin binding enzymes Biotin containing enzymes Fatty acids binding proteins Lipoproteins, lipases, coagulation factors, DNA polymerases, steroid receptor proteins, growth factors, serine protease inhibitors Immunoglobulins Glycoproteins containing -D-mannopyranosyl and -Dglucopyranosyl residues Glycoproteins containing N-acetyl -(or ) -Dgalactopyranosyl residues Glycoproteins RNA containing poly (U) sequences, some RNA specific proteins r. RNA

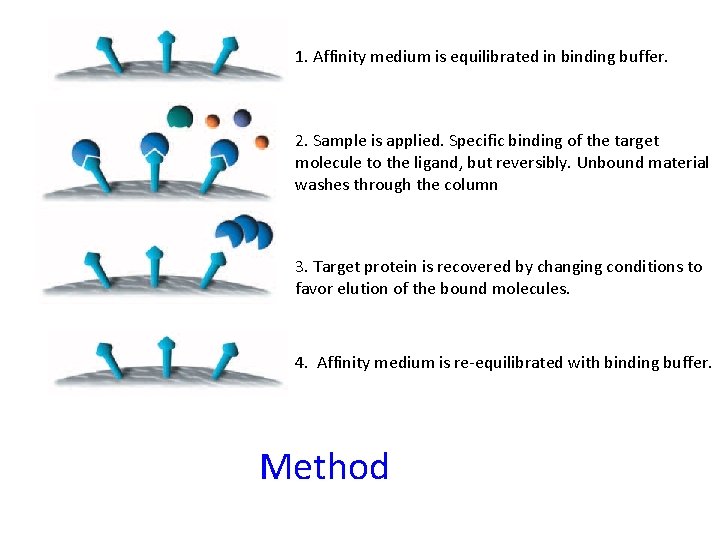
1. Affinity medium is equilibrated in binding buffer. 2. Sample is applied. Specific binding of the target molecule to the ligand, but reversibly. Unbound material washes through the column 3. Target protein is recovered by changing conditions to favor elution of the bound molecules. 4. Affinity medium is re-equilibrated with binding buffer. Method

Some typical biological interactions • Enzyme- substrate or inhibitor or cofactor • Antibody- antigen or virus or cell • Lectin – polysaccharide or glycoprotein or cell surface receptor, cell • Nucleic acid - complementary base sequence or histones or nucleic acid polymerase or nucleic acid binding protein. • Hormone/vitamin – receptor or carrier protein. • Glutathione - glutathione-S-transferase or GST fusion proteins. • Metal ions - Poly (His) fusion proteins or native proteins with histidine, cysteine, tryptophan residues on their surfaces.

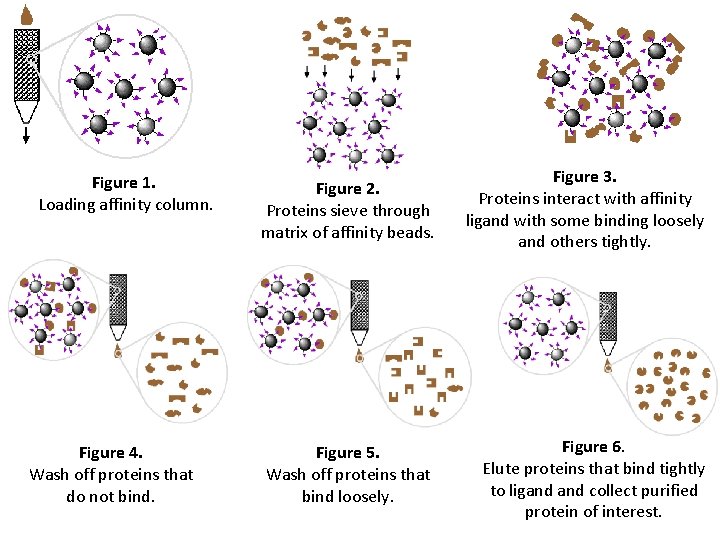
Figure 1. Loading affinity column. Figure 4. Wash off proteins that do not bind. Figure 2. Proteins sieve through matrix of affinity beads. Figure 3. Proteins interact with affinity ligand with some binding loosely and others tightly. Figure 5. Wash off proteins that bind loosely. Figure 6. Elute proteins that bind tightly to ligand collect purified protein of interest.

Applications • Purification of enzymes, immunoglobulins and receptor proteins • Isolation of nucleic acids. eg. m. RNA by selective hybridization of poly (U) – Sepharose 4 B by exploiting its poly (U) tail. • Isolation of complementary RNA and DNA using immobilized single stranded DNA. • Isolation of proteins involved in nucleic acid metabolism by immobilized nucleotides