Advancing Science with DNA Sequence Microbial Genome Annotation
Advancing Science with DNA Sequence Microbial Genome Annotation Nikos Kyrpides Genome Biology Program (GBP) DOE Joint Genome institute

Advancing Science with DNA Sequence Two main goals of genome analysis: • Evolutionary analysis – How does an organism compare to the rest? • Metabolic reconstruction – What can an organism do and how?
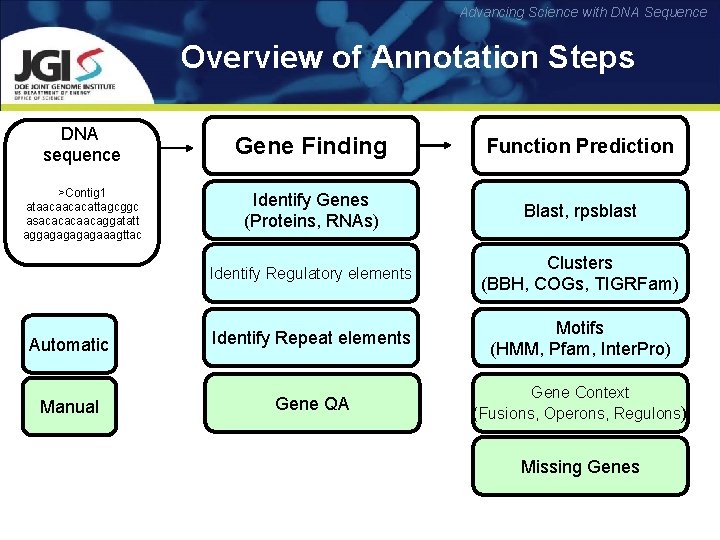
Advancing Science with DNA Sequence Overview of Annotation Steps DNA sequence Gene Finding Function Prediction >Contig 1 ataacaacacattagcggc asacacacaacaggatatt aggagagaaagttac Identify Genes (Proteins, RNAs) Blast, rpsblast Identify Regulatory elements Clusters (BBH, COGs, TIGRFam) Automatic Identify Repeat elements Motifs (HMM, Pfam, Inter. Pro) Manual Gene QA Gene Context (Fusions, Operons, Regulons) Missing Genes

Advancing Science with DNA Sequence 1. Finding the genes in microbial genomes 1. Introduction 2. Tools out there 3. Basic principles behind tools 4. Known problems of the tools: why you may need manual curation
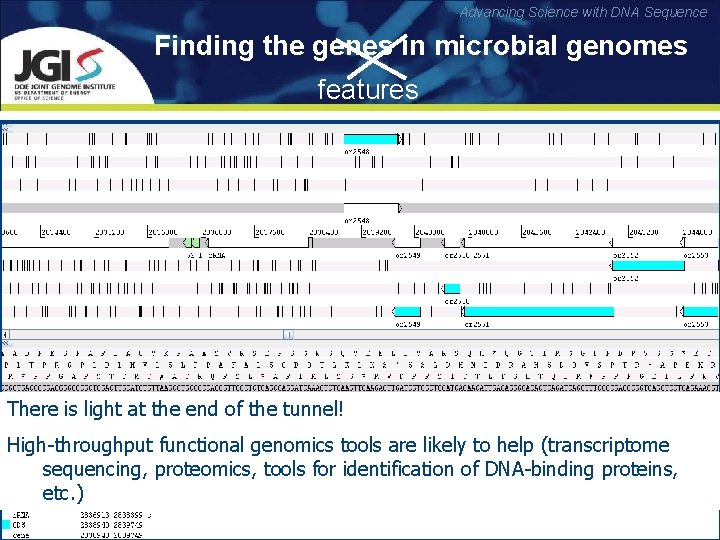
Advancing Science with DNA Sequence Finding the genes in microbial genomes features Well-annotated bacterial genome in Artemis genome viewer: Sequence features in prokaryotic genomes: § stable RNA-coding genes (r. RNAs, t. RNAs, RNA component of RNase. P, tm. RNA) § protein-coding genes (CDSs) § transcriptional features (m. RNAs, operons, promoters, terminators, protein-binding sites, DNA bends) § translational features (RBS, regulatory antisense r. RNA t. RNAs, m. RNA secondary structures, translational operon There is light at the end of the tunnel! recoding and programmed frameshifts, inteins) promoter terminator High-throughput functional genomics are likely toprotein-coding gene help genes) (transcriptome § pseudogenes (t. RNA and tools protein-coding sequencing, proteomics, tools for identification of DNA-binding proteins, CDS § …etc. ) protein-binding site

Advancing Science with DNA Sequence Finding the genes in microbial genomes 1. Introduction 2. Tools out there 3. Basic principles behind tools 4. Known problems of the tools: why you may need manual curation
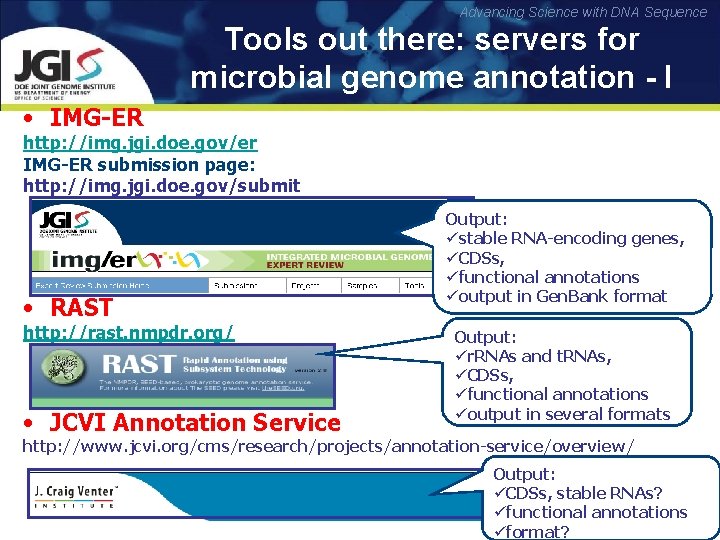
Advancing Science with DNA Sequence Tools out there: servers for microbial genome annotation - I • IMG-ER http: //img. jgi. doe. gov/er IMG-ER submission page: http: //img. jgi. doe. gov/submit • RAST http: //rast. nmpdr. org/ • JCVI Annotation Service Output: üstable RNA-encoding genes, üCDSs, üfunctional annotations üoutput in Gen. Bank format Output: ür. RNAs and t. RNAs, üCDSs, üfunctional annotations üoutput in several formats http: //www. jcvi. org/cms/research/projects/annotation-service/overview/ Output: üCDSs, stable RNAs? üfunctional annotations üformat?
Advancing Science with DNA Sequence Tools out there: servers for microbial genome annotation - II • AMIGENE http: //www. genoscope. cns. fr/agc/tools/amiga/Form/form. php • Ref. Seq Output: CDSs, output in gff format http: //www. ncbi. nlm. nih. gov/genomes/MICROBES/genemark. cgi http: //www. ncbi. nlm. nih. gov/genomes/MICROBES/glimmer_3. cgi • Easy. Gene Output: CDSs, output in tbl format http: //www. cbs. dtu. dk/services/Easy. Gene/ Output: CDSs, size restriction <1 Mb
Advancing Science with DNA Sequence Tools out there: genome browsers for manual annotation of microbial genomes • Artemis http: //www. sanger. ac. uk/Software/Artemis/ • Manatee http: //manatee. sourceforge. net/ • Argo http: //www. broad. mit. edu/annotation/argo/ Windows and Linux versions; works with files in many formats, annotated by any pipeline Linux versions only; genome needs to be annotated by the JCVI Annotation Service Windows and Linux; works with files in many formats Major difference: viewer vs editor?
Advancing Science with DNA Sequence Tools out there: tools for finding stable (“non-coding”) RNAs - I • Large structural RNAs (23 S and 16 S r. RNAs) RNAmmer http: //www. cbs. dtu. dk/services/RNAmmer/ • Small structural RNAs (5 S r. RNA, t. RNAs, tm. RNA, RNase. P RNA component) Rfam database, INFERNAL search tool http: //www. sanger. ac. uk/Software/Rfam/ http: //rfam. janelia. org/ http: //infernal. janelia. org/ Web service: sequence search is limited to 2 kb ARAGORN Web service: sequence http: //130. 235. 46. 10/ARAGORN 1. 1/HTML/aragorn 1. 2. html t. RNAScan-SE http: //lowelab. ucsc. edu/t. RNAscan-SE/ search is limited to 15 kb, finds t. RNAs and tm. RNAs only Web service: sequence search is limited to 5 Mb, finds t. RNAs only
Advancing Science with DNA Sequence Tools out there: tools for finding “non-coding” RNAs - II • Short regulatory RNAs Rfam database, INFERNAL search tool http: //www. sanger. ac. uk/Software/Rfam/ http: //rfam. janelia. org/ http: //infernal. janelia. org/ Web service: sequence search is limited to 2 kb; Provides list of precalculated RNAs for publicly available genomes Other (less popular) tools: Pipeline for discovering cis-regulatory nc. RNA motifs: http: //bio. cs. washington. edu/supplements/yzizhen/pipeline/ RNAz http: //www. tbi. univie. ac. at/~wash/RNAz/
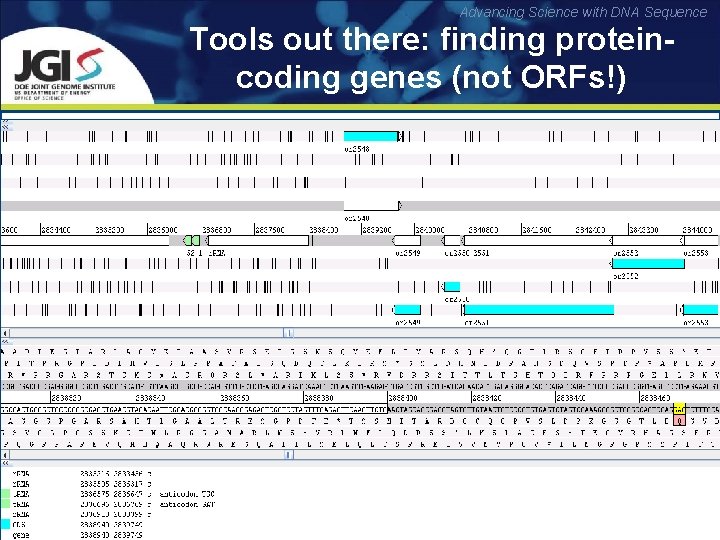
Advancing Science with DNA Sequence Tools out there: finding proteincoding genes (not ORFs!) Reading frames: translations of the nucleotide sequence with an offset of 0, 1 and 2 nucleotides (three possible translations in each direction) Open reading frame (ORF): reading frame between a start and stop codon
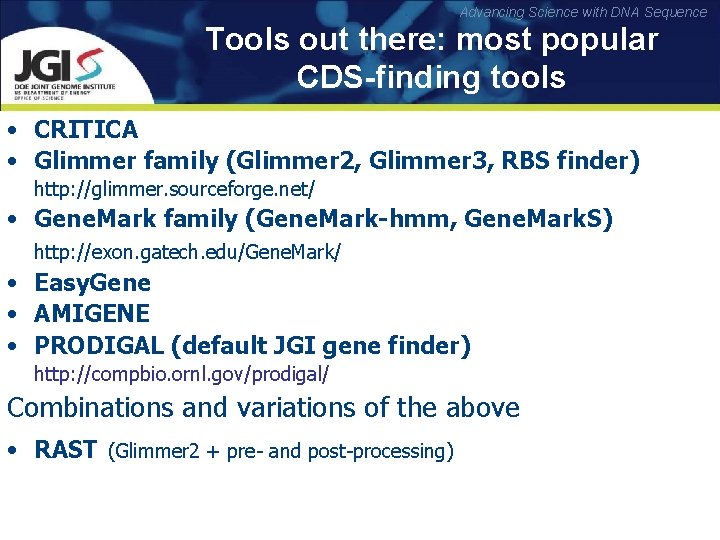
Advancing Science with DNA Sequence Tools out there: most popular CDS-finding tools • CRITICA • Glimmer family (Glimmer 2, Glimmer 3, RBS finder) http: //glimmer. sourceforge. net/ • Gene. Mark family (Gene. Mark-hmm, Gene. Mark. S) http: //exon. gatech. edu/Gene. Mark/ • Easy. Gene • AMIGENE • PRODIGAL (default JGI gene finder) http: //compbio. ornl. gov/prodigal/ Combinations and variations of the above • RAST (Glimmer 2 + pre- and post-processing)

Advancing Science with DNA Sequence Finding the genes in microbial genomes 1. Introduction 2. Tools out there 3. Basic principles behind tools (very basic, see specific papers for details) 4. Known problems of the tools: why you may need manual curation
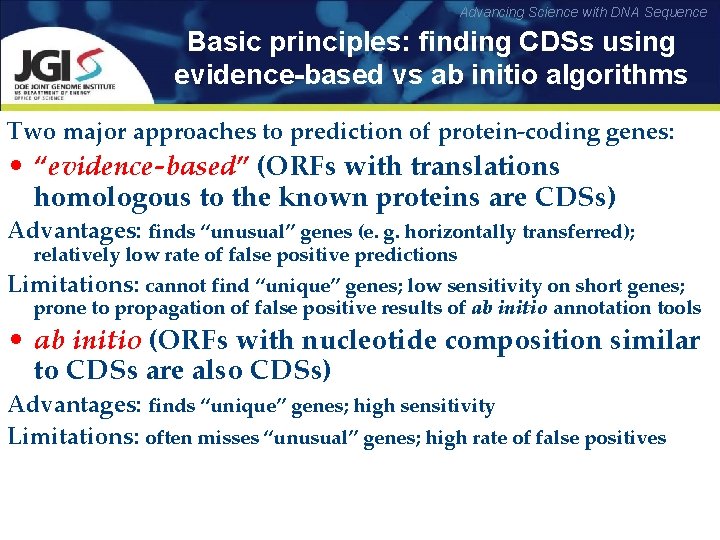
Advancing Science with DNA Sequence Basic principles: finding CDSs using evidence-based vs ab initio algorithms Two major approaches to prediction of protein-coding genes: • “evidence-based” (ORFs with translations homologous to the known proteins are CDSs) Advantages: finds “unusual” genes (e. g. horizontally transferred); relatively low rate of false positive predictions Limitations: cannot find “unique” genes; low sensitivity on short genes; prone to propagation of false positive results of ab initio annotation tools • ab initio (ORFs with nucleotide composition similar to CDSs are also CDSs) Advantages: finds “unique” genes; high sensitivity Limitations: often misses “unusual” genes; high rate of false positives

Advancing Science with DNA Sequence Finding the genes in microbial genomes 1. Introduction 2. Tools out there 3. Basic principles behind tools 4. Known problems of the tools: why you may need manual curation

Advancing Science with DNA Sequence Known problems: CDSs § ü ü Short CDSs: many are missed, others are overpredicted short ribosomal proteins (30 -40 aa long) are often missed short proteins in the promoter region are often overpredicted N-terminal sequences are often inaccurate (many features of the sequence around start codon are not accounted for) Glimmer 2. 0 is calling genes longer than they should be Gene. Mark, Glimmer 3. 0 mostly call genes shorter Pseudogenes and sequencing errors (artificial frameshift) all tools are looking for ORFs (needs valid start and stop codons) “unique” genes are often predicted on the opposite strand of a pseudogene or a gene with a sequencing error Proteins with unusual translational features (recoding, programmed frameshifts) these genes are often mistaken for pseudogenes see pseudogenes

Advancing Science with DNA Sequence Known problems: CDSs Lack of Standards
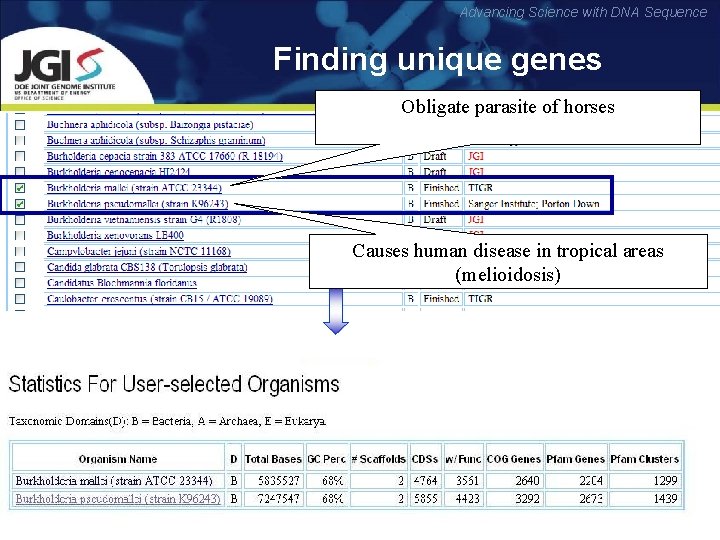
Advancing Science with DNA Sequence Finding unique genes Obligate parasite of horses Causes human disease in tropical areas (melioidosis)
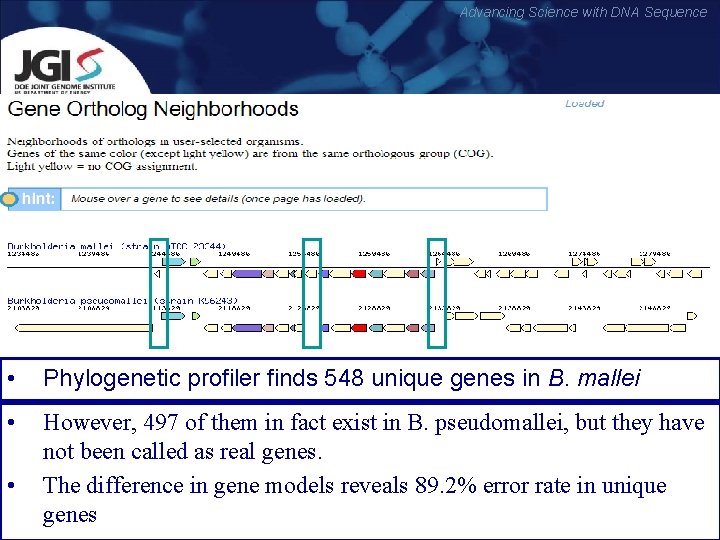
Advancing Science with DNA Sequence • Phylogenetic profiler finds 548 unique genes in B. mallei • However, 497 of them in fact exist in B. pseudomallei, but they have not been called as real genes. The difference in gene models reveals 89. 2% error rate in unique genes •
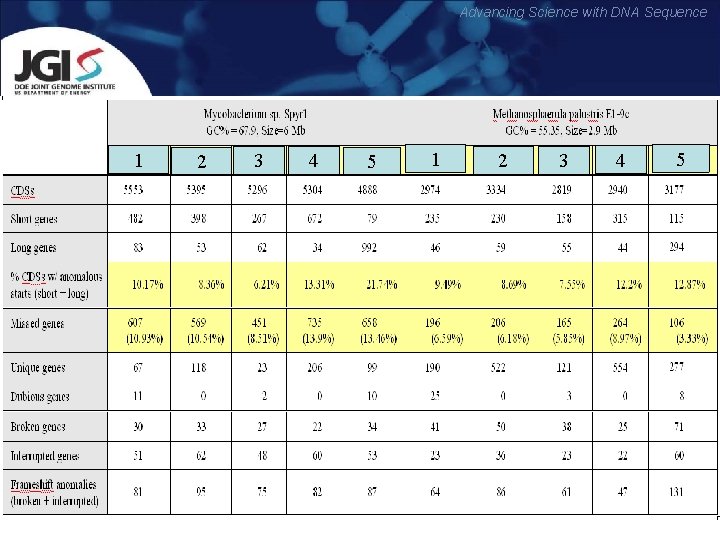
Advancing Science with DNA Sequence 1 2 3 4 5
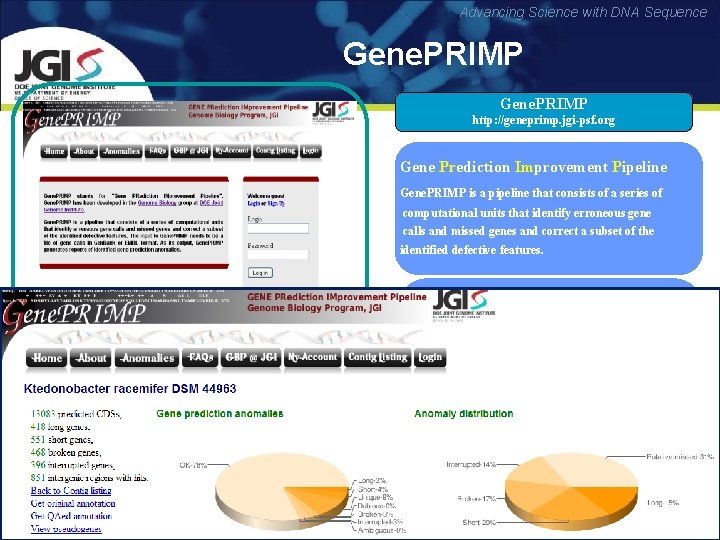
Advancing Science with DNA Sequence Gene. PRIMP http: //geneprimp. jgi-psf. org Gene Prediction Improvement Pipeline Pr Im Gene. PRIMP is a pipeline that consists of a series of computational units that identify erroneous gene calls and missed genes and correct a subset of the identified defective features. APPLICATIONS • Identify gene prediction anomalies • Benchmark the quality of gene prediction algorithms • Benchmark the quality of combination / coverage of sequencing platforms • Improve the sequence quality Pati A. et al, Nature Methods June 2010
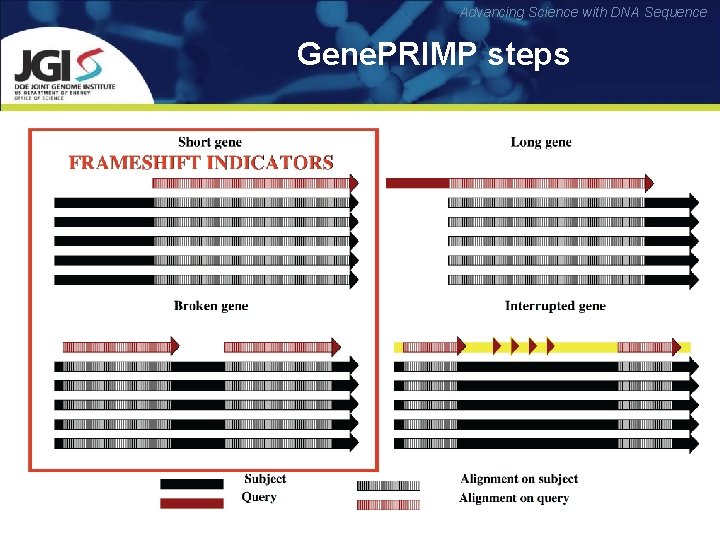
Advancing Science with DNA Sequence Gene. PRIMP steps
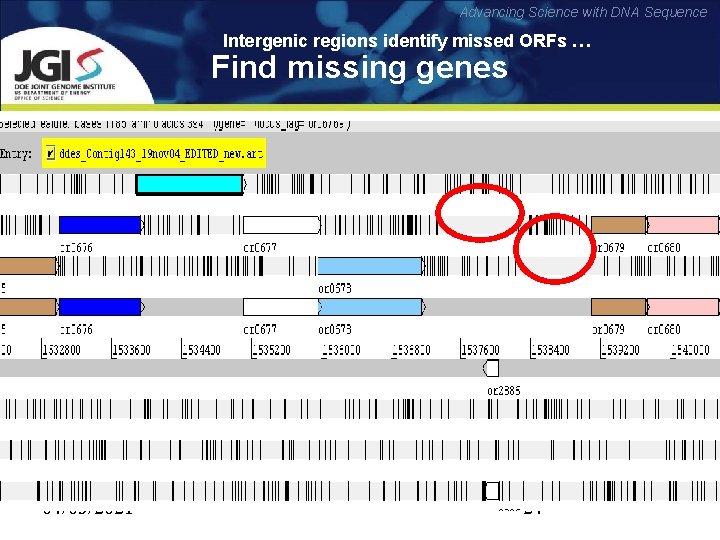
Advancing Science with DNA Sequence Intergenic regions identify missed ORFs … Find missing genes 04/03/2021 24
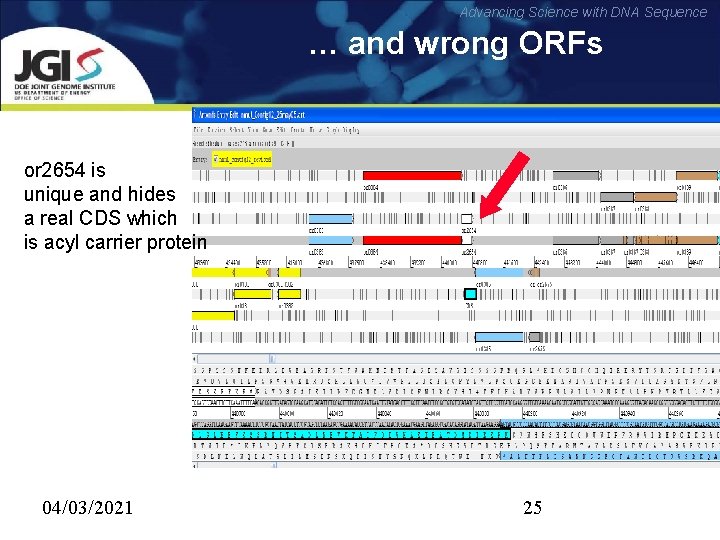
Advancing Science with DNA Sequence … and wrong ORFs or 2654 is unique and hides a real CDS which is acyl carrier protein 04/03/2021 25
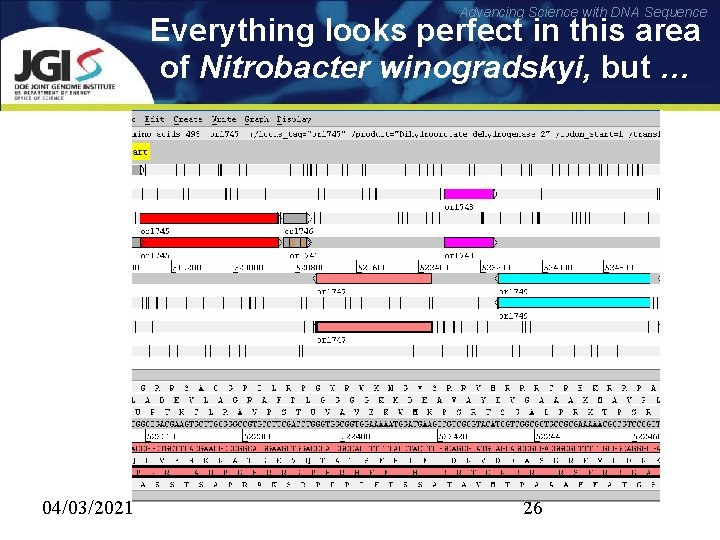
Advancing Science with DNA Sequence Everything looks perfect in this area of Nitrobacter winogradskyi, but … 04/03/2021 26

Advancing Science with DNA Sequence … hides a real ORF 04/03/2021 27

Guinness Book of protein-coding genes Advancing Science with DNA Sequence ü The longest human gene is 2, 220, 223 nucleotides long. It has 79 exons, with a total of only 11, 058 nucleotides, which specify the sequence of the 3, 685 amino acids and codes for a protein dystrophin. It is part of a protein complex located in the cell membrane, which transfers the force generated by the actinmyosin structure inside the muscle fiber to the entire fiber. ü The smallest human gene is 252 nucleotides long, it specifies a polypeptide of 67 amino acids and codes for an insulin-like growth factor II. ü The longest bacterial gene is 110, 418 nucleotides long, which specify the sequence of 36, 805 amino acids. Its function is unknown, most likely a surface protein. ü The smallest bacterial gene is 54 nucleotides long, it specifies a polypeptide of 17 amino acids and codes for a regulatory protein in cyanobacteria

Advancing Science with DNA Sequence 1. Finding the functions in microbial genomes 1. Introduction 2. Tools out there 3. Known problems
Advancing Science with DNA Sequence Functional characterization
Advancing Science with DNA Sequence Computational approaches to Functional characterization
Advancing Science with DNA Sequence Homology Two sequences are homologous, if there existed a molecule in the past that is ancestral to both of the sequences. Types of Homology: üOrthology: bifurcation in molecular tree reflects speciation üParalogy: bifurcation in molecular tree reflects gene duplication üXenology: gene was obtained by organism through horizontal transfer üSynology: genes ended up in one organism through fusion of lineages

Advancing Science with DNA Sequence Homology & analogy • The term homology is confounded & abused in the literature! – sequences are homologous if they’re related by divergence from a common ancestor – analogy relates to the acquisition of common features from unrelated ancestors via convergent evolution • e. g. , b-barrels occur in soluble serine proteases & integral membrane porins; chymotrypsin & subtilisin share groups of catalytic residues, with near identical spatial geometries, but no other similarities • Homology is not a measure of similarity & is not quantifiable – it is an absolute statement that sequences have a divergent rather than a convergent relationship – the phrases "the level of homology is high" or "the sequences show 50% homology", or any like them, are strictly meaningless!
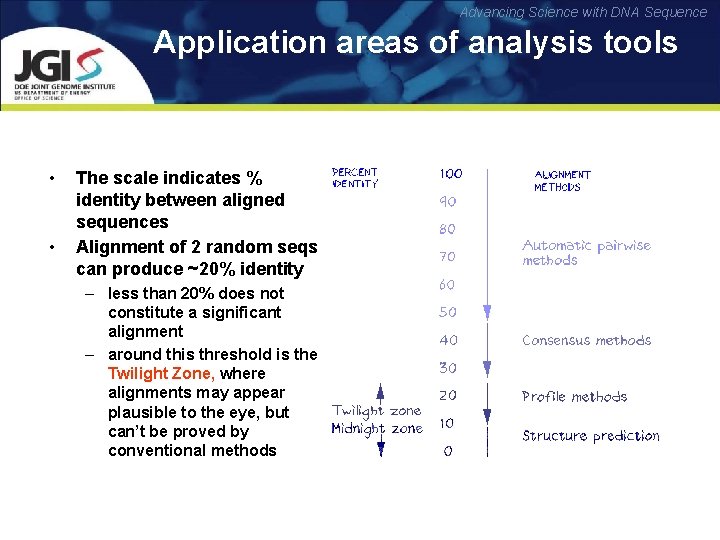
Advancing Science with DNA Sequence Application areas of analysis tools • • The scale indicates % identity between aligned sequences Alignment of 2 random seqs can produce ~20% identity – less than 20% does not constitute a significant alignment – around this threshold is the Twilight Zone, where alignments may appear plausible to the eye, but can’t be proved by conventional methods

Advancing Science with DNA Sequence Finding the functions in microbial genomes 1. Introduction 2. Tools out there 3. Known problems
Advancing Science with DNA Sequence Function prediction • Similarity searches (BLAST). • Domain identification(Pfam). • Small sequence identification(PROSITE). • Localization (membrane, intracellular, extracellular). • Other sequence features. • Similarity to known structures.
Advancing Science with DNA Sequence Primary nucleotide/protein databases EMBL/Gen. Bank/DDBJ (http: //www. ncbi. nlm. nih. gov/ , http: //www. ebi. ac. uk/embl ) Archive containing all sequences from: genome projects sequencing centers individual scientists patent offices The sequences are exchanged between the three centers on a daily basis. Database is doubling every 10 months. Sequences from >140, 000 different species. 1400 new species added every month. Database name nt / nr

Advancing Science with DNA Sequence Ref. Seq Curated transcripts and proteins. reviewed by NCBI staff. Model transcripts and proteins. generated by computer algorithms. Assembled Genomic Regions (contigs). Chromosome records.
Advancing Science with DNA Sequence Classification databases Groups (families/clusters) of proteins based on… Overall sequence similarity. Local sequence similarity. Presence / absence of specific features. Structural similarity. . These groups contain proteins with similar properties. Specific function, enzymatic activity. Broad function. Evolutionary relationship. …
Advancing Science with DNA Sequence Clusters of orthologous genes (COGs) COGs were delineated by comparing protein sequences encoded in 43 complete genomes representing 30 major phylogenetic lineages. Each Cluster has representatives of at least 3 lineages A function (specific or broad) has been assigned to each COG. Functions grouped to functional categories. COGNITOR reverse psi BLAST (CDD) http: //www. ncbi. nlm. nih. gov/COG/
Advancing Science with DNA Sequence Profiles & Pfam • A method for classifying proteins into groups exploits region similarities, which contain valuable information (domains/profiles). • These domains/profiles can be used to detect distant relationships, where only few residues are conserved.
Pfam http: //pfam. sanger. ac. uk Advancing Science with DNA Sequence
![Advancing Science with DNA Sequence PROSITE http: //au. expasy. org/prosite/ R-Y-x-[DT]-W-x-[LIVM]-[ST]-T-P-[LIVM](3) Advancing Science with DNA Sequence PROSITE http: //au. expasy. org/prosite/ R-Y-x-[DT]-W-x-[LIVM]-[ST]-T-P-[LIVM](3)](http://slidetodoc.com/presentation_image_h/22916ff8f4c3069710ea3987a796e58c/image-43.jpg)
Advancing Science with DNA Sequence PROSITE http: //au. expasy. org/prosite/ R-Y-x-[DT]-W-x-[LIVM]-[ST]-T-P-[LIVM](3)
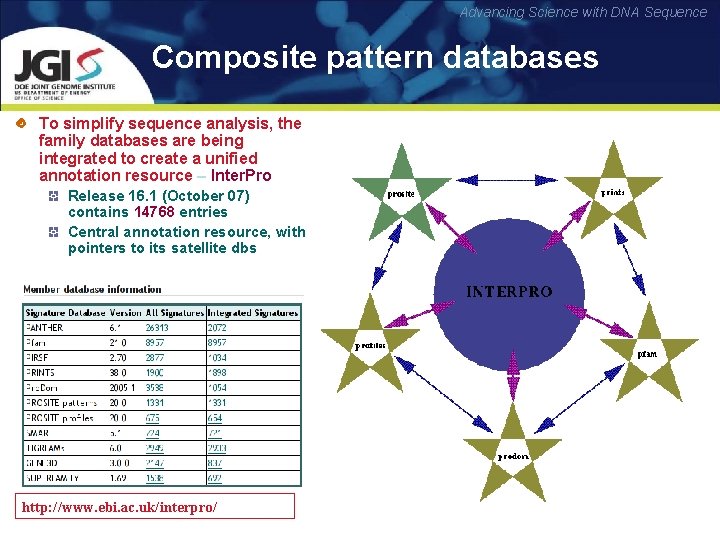
Advancing Science with DNA Sequence Composite pattern databases To simplify sequence analysis, the family databases are being integrated to create a unified annotation resource – Inter. Pro Release 16. 1 (October 07) contains 14768 entries Central annotation resource, with pointers to its satellite dbs http: //www. ebi. ac. uk/interpro/
Advancing Science with DNA Sequence * It is up to the user to decide if the annotation is correct *
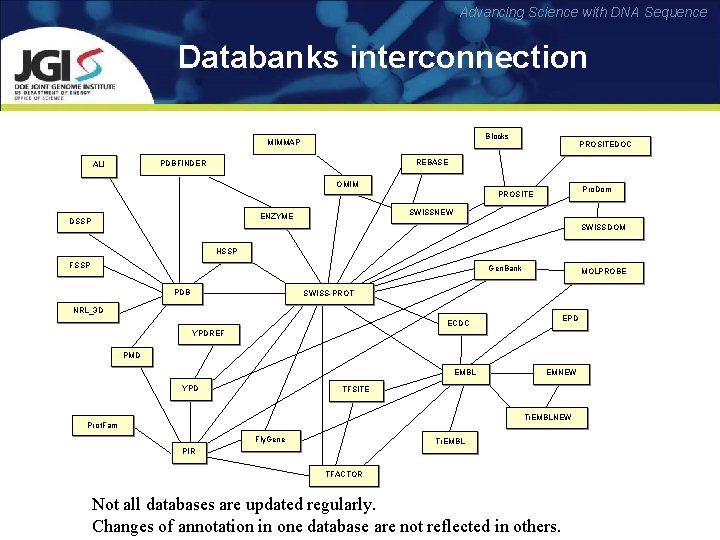
Advancing Science with DNA Sequence Databanks interconnection Blocks MIMMAP REBASE PDBFINDER ALI PROSITEDOC OMIM Pro. Dom PROSITE SWISSNEW ENZYME DSSP SWISSDOM HSSP FSSP Gen. Bank PDB MOLPROBE SWISS-PROT NRL_3 D EPD ECDC YPDREF PMD EMBL YPD EMNEW TFSITE Tr. EMBLNEW Prot. Fam Fly. Gene Tr. EMBL PIR TFACTOR Not all databases are updated regularly. Changes of annotation in one database are not reflected in others.
Advancing Science with DNA Sequence Summary • We have main archives (Genbank), and currated databases (Refseq, Swiss. Prot), and protein classification database (COG, Pfam). • This is the tip of the iceberg of databases. • They help predict the function, or the network of functions. • Systems that integrate the information from several databases, visualize and allow handling of data in an intuitive way are required

Advancing Science with DNA Sequence Finding the functions in microbial genomes 1. Introduction 2. Tools out there 3. Known problems
Advancing Science with DNA Sequence Finding the functions in microbial genomes Some Problems in Function Prediction ü Quality of Public Databases ü Domain shuffling ü Orthology / Paralogy ü Horizontal gene transfers ü Hypothetical proteins – Unknown genes
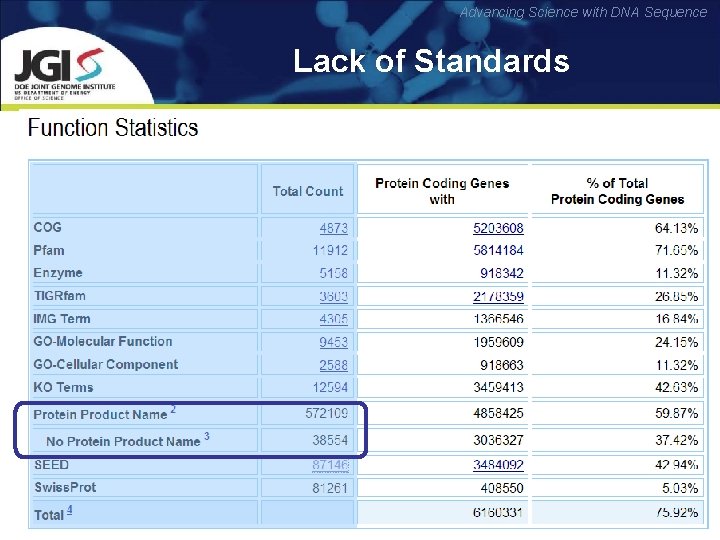
Advancing Science with DNA Sequence Lack of Standards
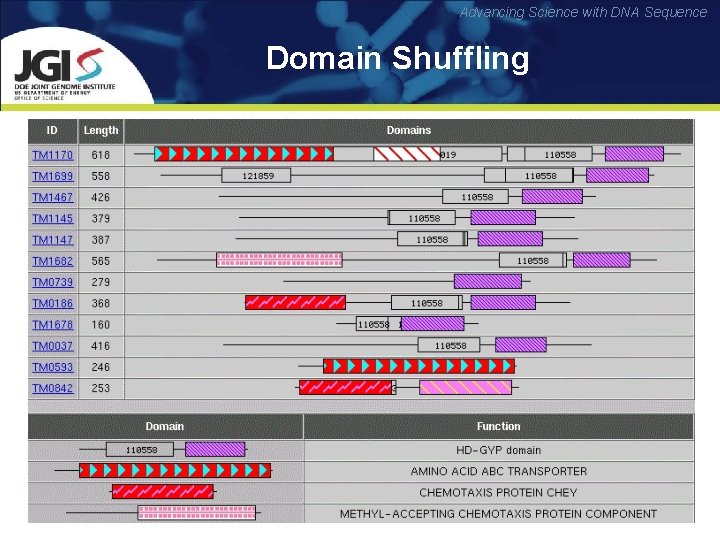
Advancing Science with DNA Sequence Domain Shuffling
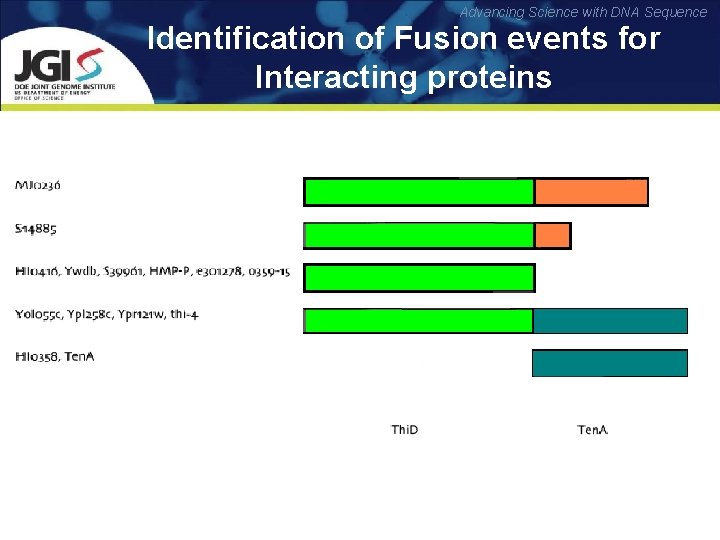
Advancing Science with DNA Sequence Identification of Fusion events for Interacting proteins

Advancing Science with DNA Sequence Ortholog – Paralog muddle
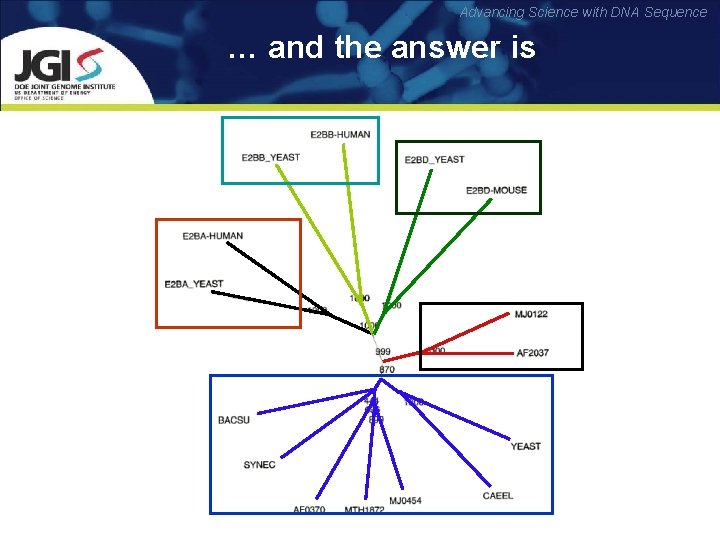
Advancing Science with DNA Sequence … and the answer is
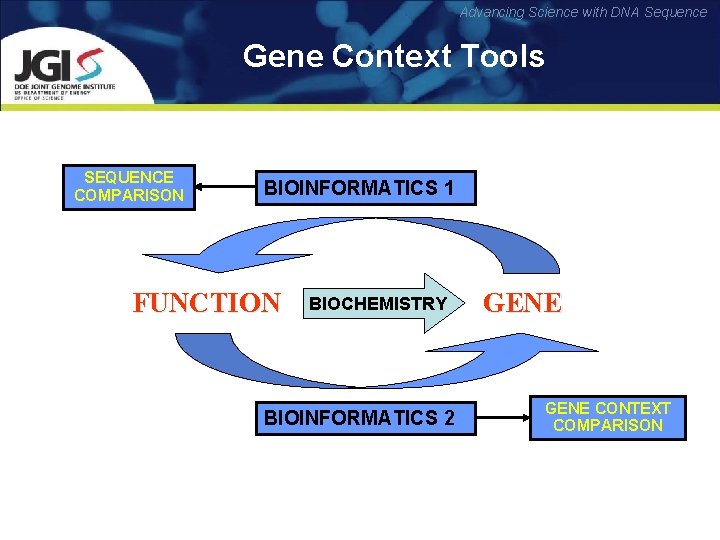
Advancing Science with DNA Sequence Gene Context Tools SEQUENCE COMPARISON BIOINFORMATICS 1 FUNCTION BIOCHEMISTRY BIOINFORMATICS 2 GENE CONTEXT COMPARISON
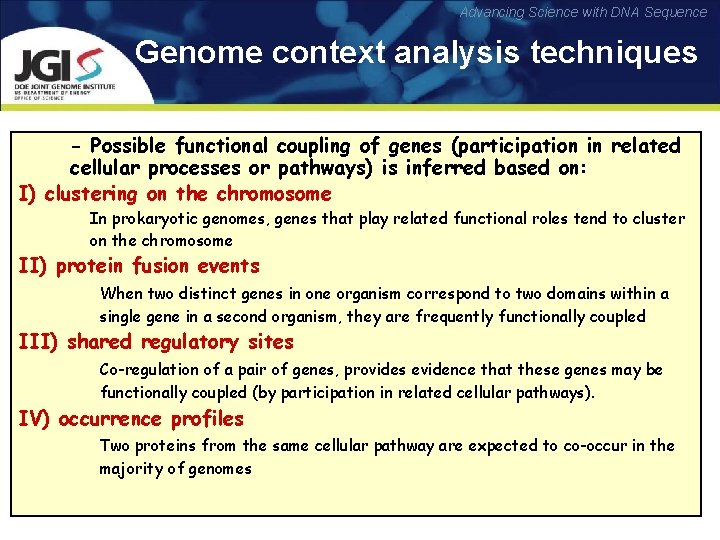
Advancing Science with DNA Sequence Genome context analysis techniques - Possible functional coupling of genes (participation in related cellular processes or pathways) is inferred based on: I) clustering on the chromosome In prokaryotic genomes, genes that play related functional roles tend to cluster on the chromosome II) protein fusion events When two distinct genes in one organism correspond to two domains within a single gene in a second organism, they are frequently functionally coupled III) shared regulatory sites Co-regulation of a pair of genes, provides evidence that these genes may be functionally coupled (by participation in related cellular pathways). IV) occurrence profiles Two proteins from the same cellular pathway are expected to co-occur in the majority of genomes
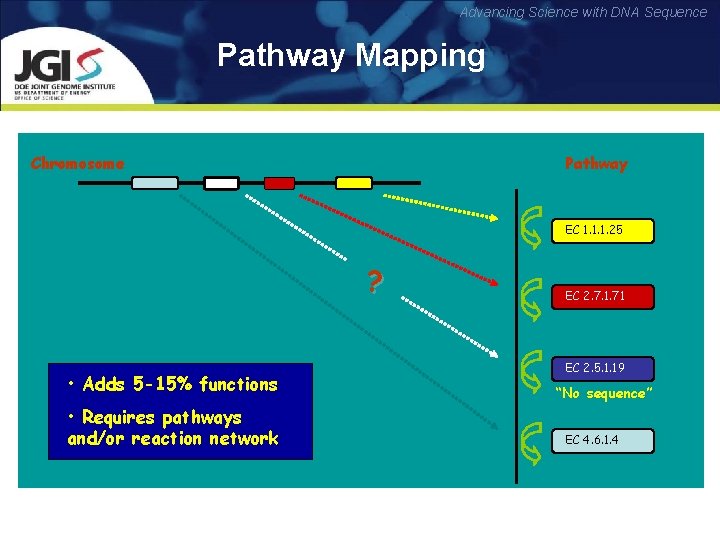
Advancing Science with DNA Sequence Pathway Mapping Chromosome Pathway EC 1. 1. 1. 25 ? • Adds 5 -15% functions • Requires pathways and/or reaction network EC 2. 7. 1. 71 EC 2. 5. 1. 19 “No sequence” EC 4. 6. 1. 4
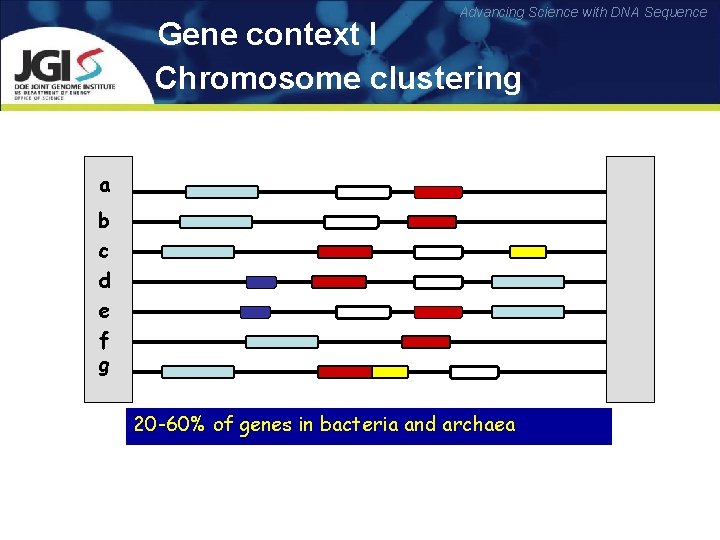
Advancing Science with DNA Sequence Gene context I Chromosome clustering Genome a b c d e f g 20 -60% of genes in bacteria and archaea
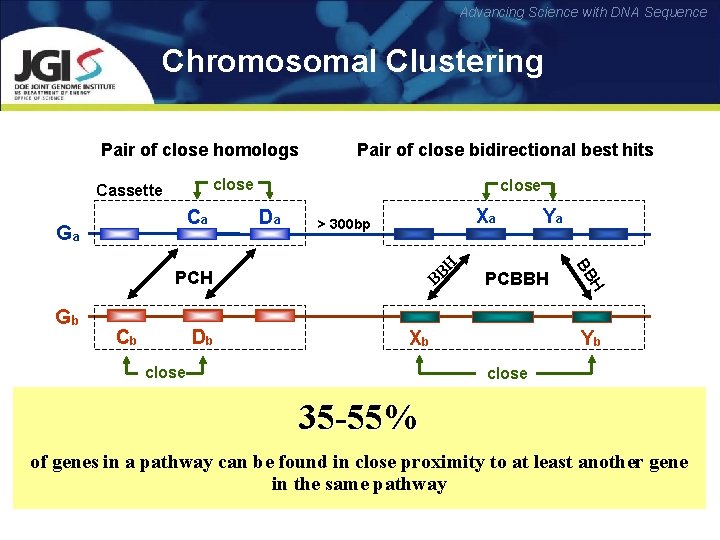
Advancing Science with DNA Sequence Chromosomal Clustering Pair of close homologs Pair of close bidirectional best hits close Cassette Ca Ga close Da Xa > 300 bp Cb Db Xb close H Gb PCBBH BB H B B PCH Ya Yb close 35 -55% of genes in a pathway can be found in close proximity to at least another gene in the same pathway
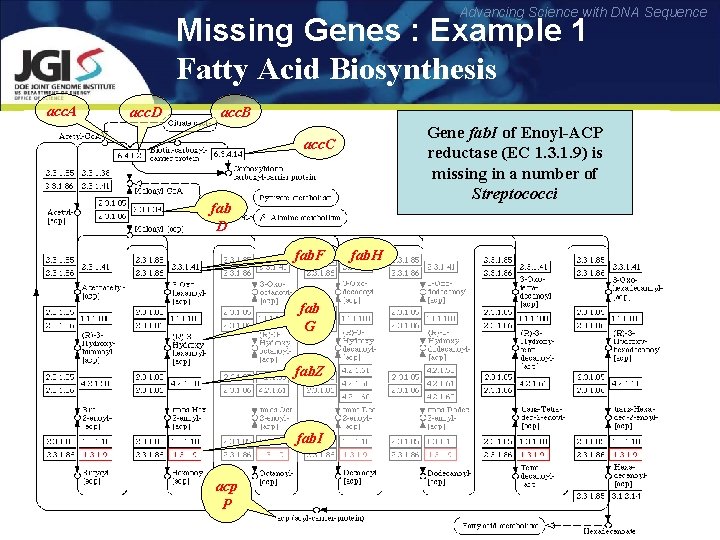
Advancing Science with DNA Sequence Missing Genes : Example 1 Fatty Acid Biosynthesis acc. A acc. D acc. B Gene fab. I of Enoyl-ACP reductase (EC 1. 3. 1. 9) is missing in a number of Streptococci acc. C fab D fab. F fab G fab. Z fab. I acp P fab. H
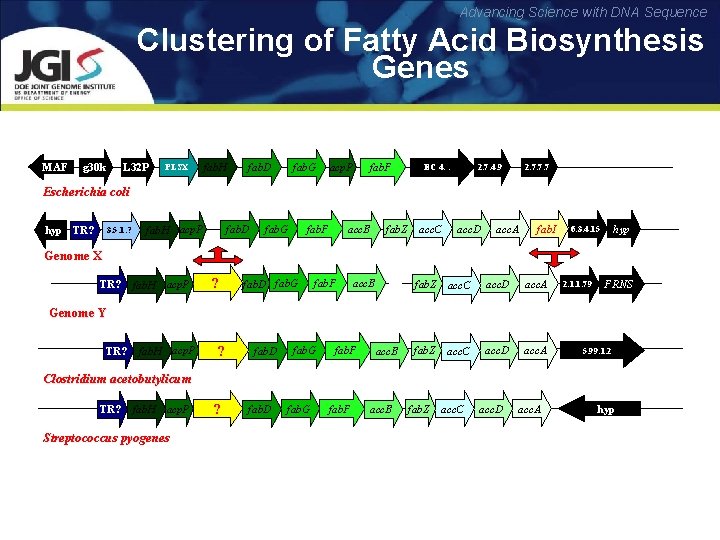
Advancing Science with DNA Sequence Clustering of Fatty Acid Biosynthesis Genes MAF g 30 k L 32 P PLSX fab. H fab. D fab. G acp. P fab. F EC 4… 2. 7. 4. 9 2. 7. 7. 7 Escherichia coli hyp TR? 3. 5. 1. ? fab. D fab. H acp. P fab. G fab. F acc. B fab. Z acc. C acc. D acc. A fab. I hyp 6. 3. 4. 15 Genome X TR? fab. H acp. P ? fab. D fab. G fab. F acc. B fab. Z acc. C acc. D acc. A 2. 1. 1. 79 FRNS Genome Y TR? fab. H acp. P ? fab. D fab. G fab. F acc. B 5. 99. 1. 2 Clostridium acetobutylicum TR? fab. H acp. P Streptococcus pyogenes ? fab. D fab. G fab. F acc. B fab. Z acc. C acc. D acc. A hyp

Advancing Science with DNA Sequence July 2000 Nature 406, 145 - 146 (2000) © Macmillan Publishers Ltd. Microbiology: A triclosan-resistant bacterial enzyme RICHARD J. HEATH AND CHARLES O. ROCK Triclosan is an antimicrobial agent that is widely used in a variety of consumer products and acts by inhibiting one of the highly conserved enzymes (enoyl-ACP reductase, or Fab. I) of bacterial fatty-acid biosynthesis. But several key pathogenic bacteria do not possess Fab. I, and here we describe a unique triclosanresistant flavoprotein, Fab. K, that can also catalyse this reaction in Streptococcus pneumoniae. Our finding has implications for the development of Fab. I-specific inhibitors as antibacterial agents.

Advancing Science with DNA Sequence Missing Genes : Example 2 Prediction and Experimental Verification of A Novel Shikimate Kinase “Operons” vs Psi-Blast
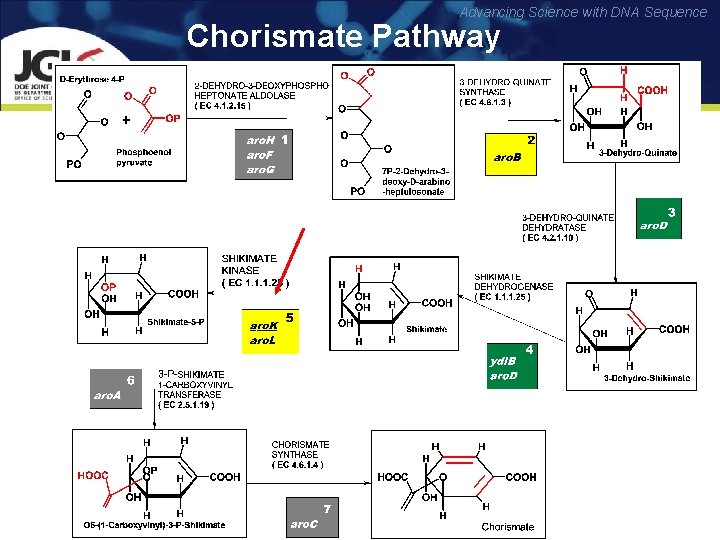
Advancing Science with DNA Sequence Chorismate Pathway
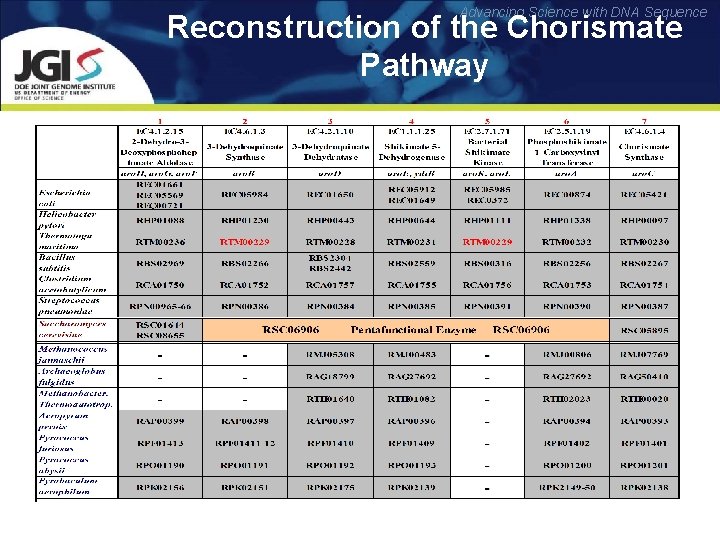
Advancing Science with DNA Sequence Reconstruction of the Chorismate Pathway
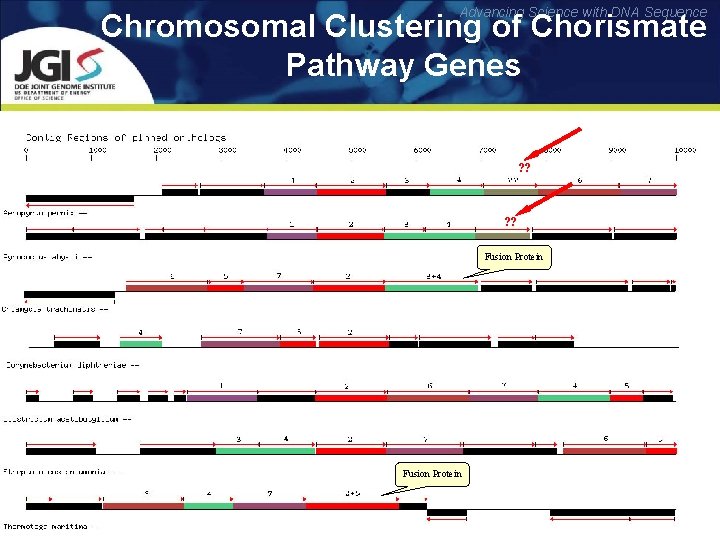
Advancing Science with DNA Sequence Chromosomal Clustering of Chorismate Pathway Genes ? ? Fusion Protein
Advancing Science with DNA Sequence Boehringer Map: an Approximation of the Core Machinery A B C D E Biochemical Pathways 1 F G H I J K L 2 3 4 5 6 7 8 9 10
Advancing Science with DNA Sequence Distribution of Universal and Local Functions (Enzymes) A B C D E Biochemical Pathways 1 F G H I J K L 2 3 4 5 6 7 8 9 10
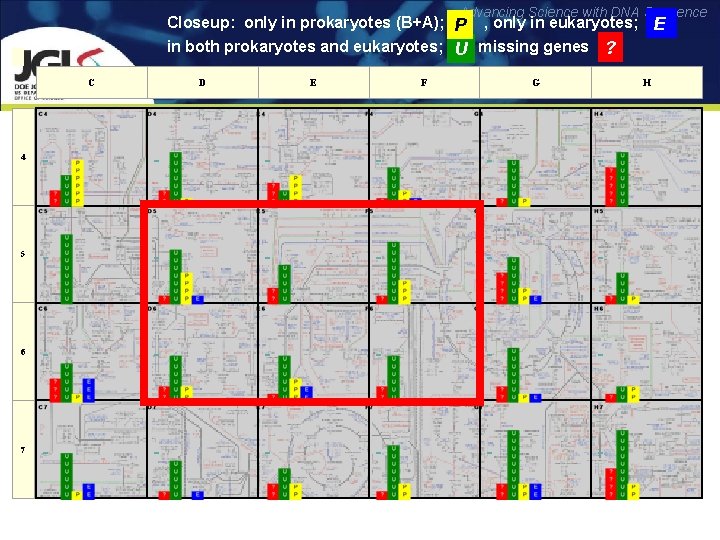
Advancing Science with DNA Sequence Closeup: only in prokaryotes (B+A); P , only in eukaryotes; E in both prokaryotes and eukaryotes; U missing genes ? C D E F G H 4 5 6 7
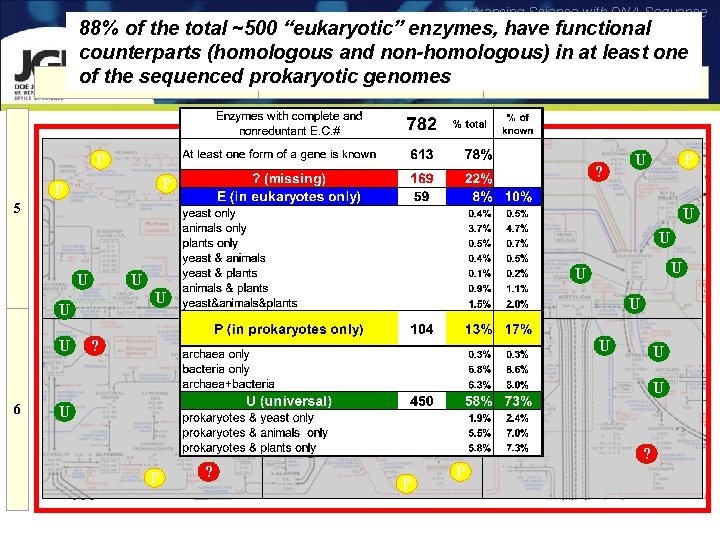
Advancing Science with DNA Sequence 88% of the total ~500 “eukaryotic” enzymes, have functional counterparts (homologous and non-homologous) in at least one of the sequenced prokaryotic genomes D E F P U U U P P U ? U U P U U U ? P 6 U U U U ? U U U U P E E U U U U U P U P ? U U U ? U P 5 ? U ? ? P P
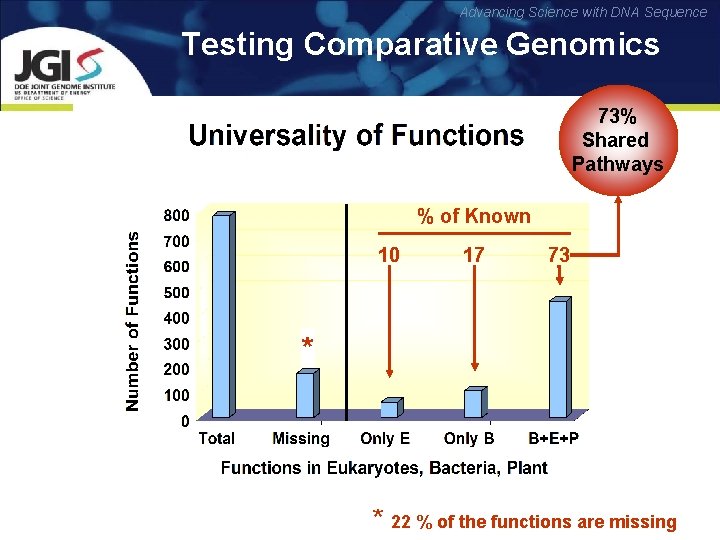
Advancing Science with DNA Sequence Testing Comparative Genomics 73% Shared Pathways % of Known 10 17 73 * * 22 % of the functions are missing
- Slides: 71