Addressable Bacterial Conjugation UC Berkeley i GEM 2006

Addressable Bacterial Conjugation UC Berkeley i. GEM 2006 Bryan Hernandez Matt Fleming Kaitlin A. Davis Jennifer Lu Samantha Liang Daniel Kluesing Will Bosworth Advisors: Professors Adam Arkin and Jay Keasling GSIs: Chris Anderson and John Dueber 1

Project Goal To establish specific cell-to-cell communication within a network of bacteria 2
. . . and make a bacterial brain 3
Project Goal F R 4
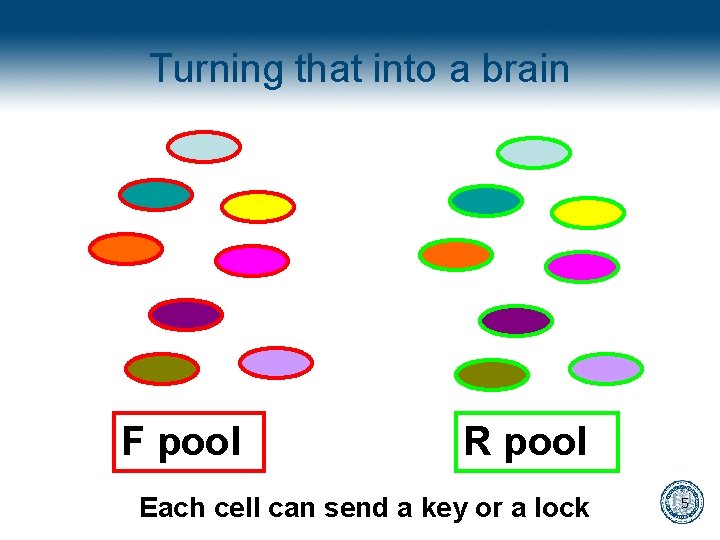
Turning that into a brain F pool R pool Each cell can send a key or a lock 5
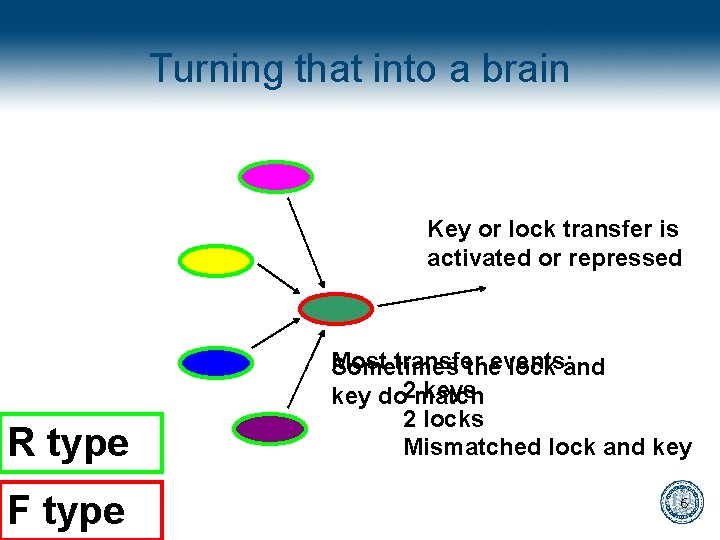
Turning that into a brain Key or lock transfer is activated or repressed R type F type Most transfer Sometimes theevents: lock and keys key do 2 match 2 locks Mismatched lock and key 6
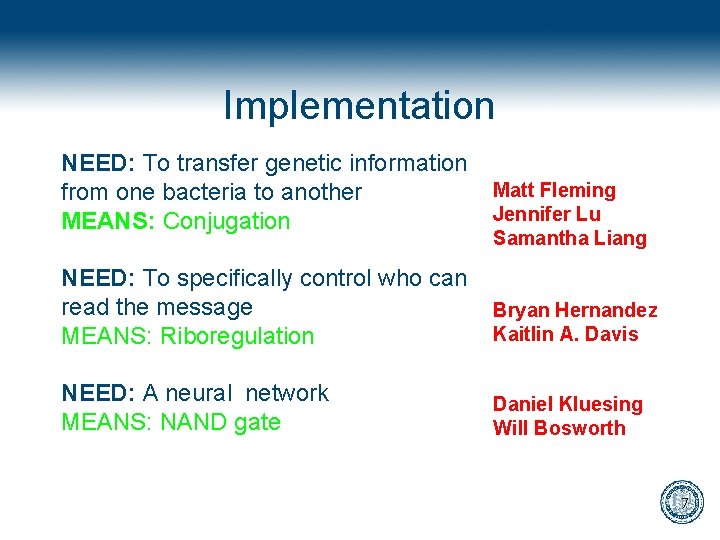
Implementation NEED: To transfer genetic information from one bacteria to another MEANS: Conjugation Matt Fleming Jennifer Lu Samantha Liang NEED: To specifically control who can read the message MEANS: Riboregulation Bryan Hernandez Kaitlin A. Davis NEED: A neural network MEANS: NAND gate Daniel Kluesing Will Bosworth 7

Conjugation Team 8
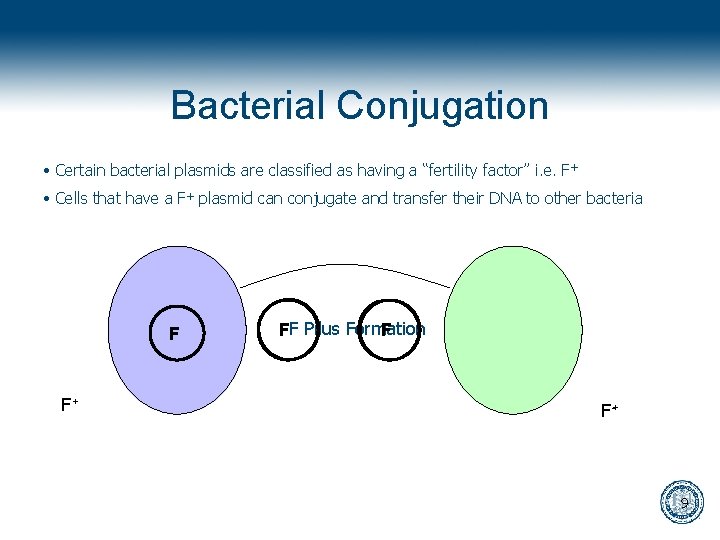
Bacterial Conjugation • Certain bacterial plasmids are classified as having a “fertility factor” i. e. F + • Cells that have a F+ plasmid can conjugate and transfer their DNA to other bacteria F F+ FF Pilus Formation F F+- 9
Relavent Information • Conjugative plasmids are very large, from 60 k – 100 k basepairs long • Many trans-acting genes are involved in the process • DNA transfer begins at a specific sequence on the plasmid, Ori. T, the Origin of Transfer. 10
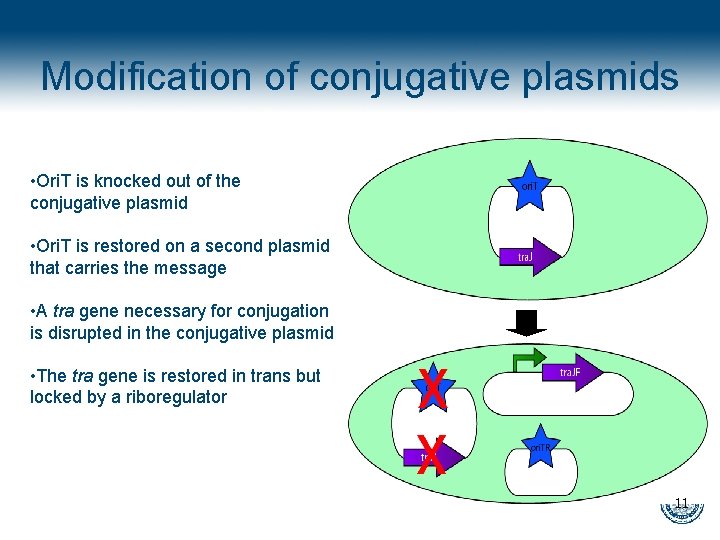
Modification of conjugative plasmids • Ori. T is knocked out of the conjugative plasmid • Ori. T is restored on a second plasmid that carries the message • A tra gene necessary for conjugation is disrupted in the conjugative plasmid • The tra gene is restored in trans but locked by a riboregulator 11
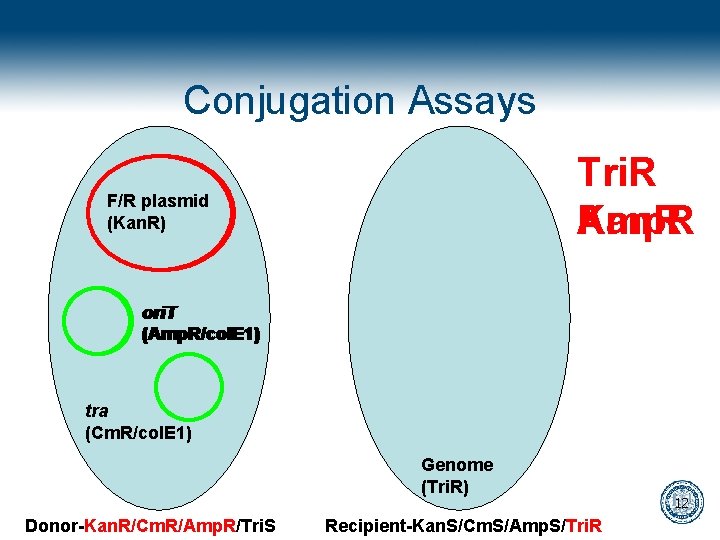
Conjugation Assays Tri. R Kan. R Amp. R F/R plasmid (Kan. R) ori. T (Amp. R/col. E 1) tra (Cm. R/col. E 1) Genome (Tri. R) Donor-Kan. R/Cm. R/Amp. R/Tri. S Recipient-Kan. S/Cm. S/Amp. S/Tri. R 12
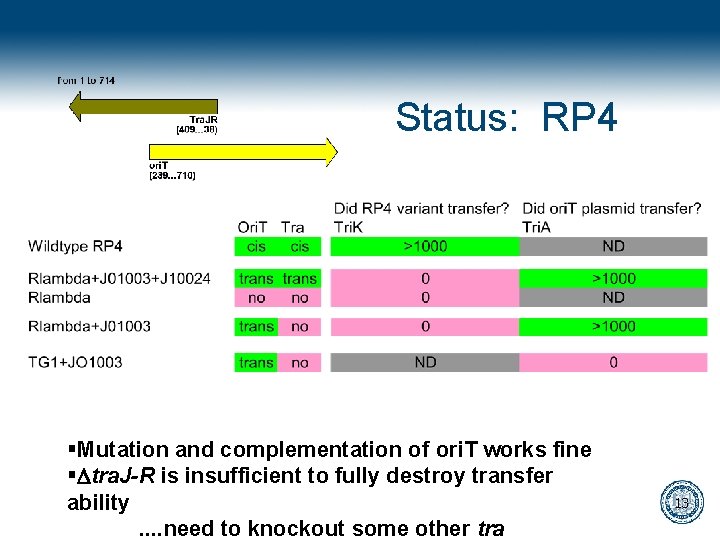
Status: RP 4 §Mutation and complementation of ori. T works fine §Dtra. J-R is insufficient to fully destroy transfer ability. . need to knockout some other tra 13
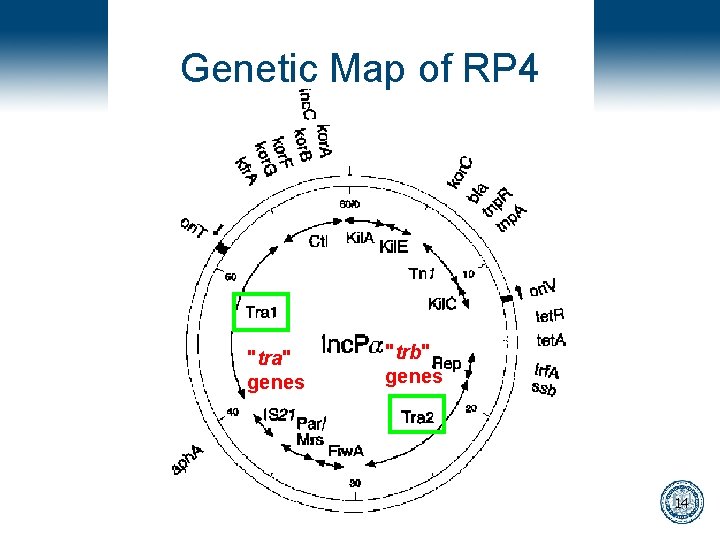
Genetic Map of RP 4 "tra" genes "trb" genes 14
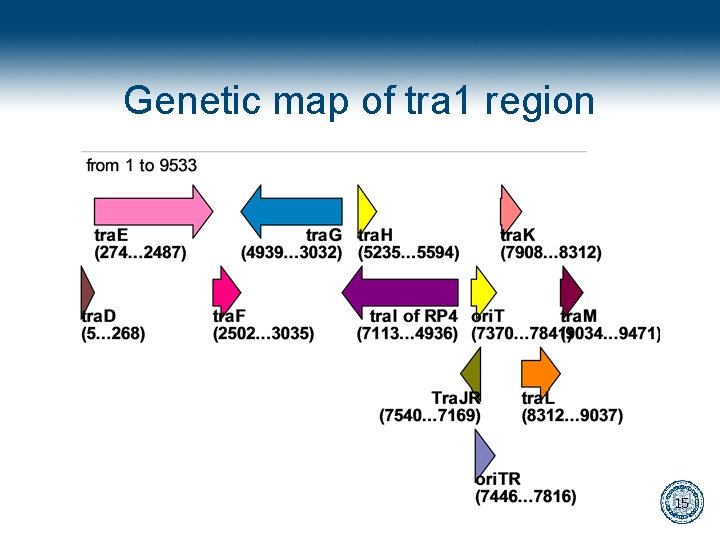
Genetic map of tra 1 region 15
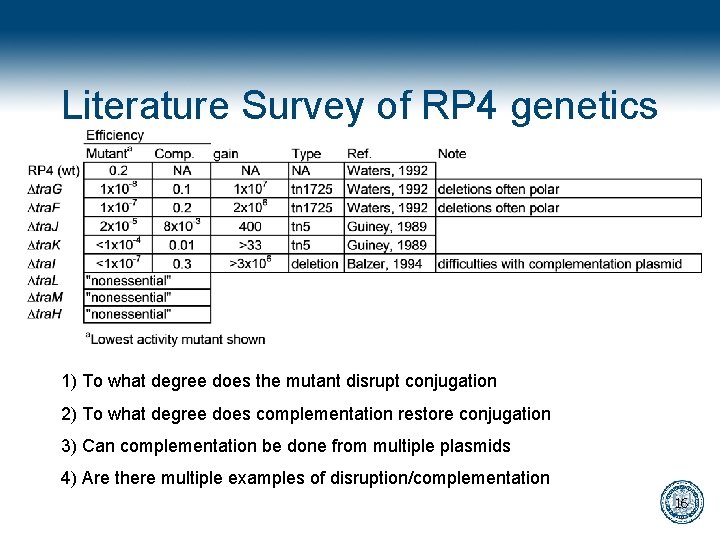
Literature Survey of RP 4 genetics 1) To what degree does the mutant disrupt conjugation 2) To what degree does complementation restore conjugation 3) Can complementation be done from multiple plasmids 4) Are there multiple examples of disruption/complementation 16
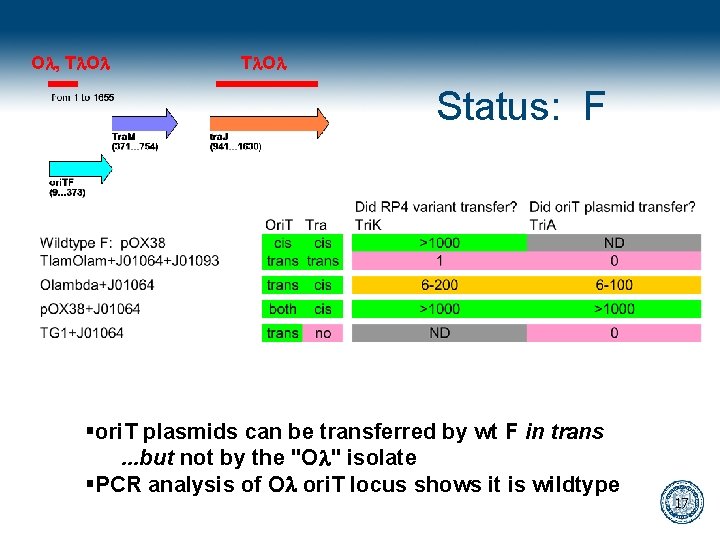
Ol, Tl. Ol Status: F §ori. T plasmids can be transferred by wt F in trans. . . but not by the "Ol" isolate §PCR analysis of Ol ori. T locus shows it is wildtype 17
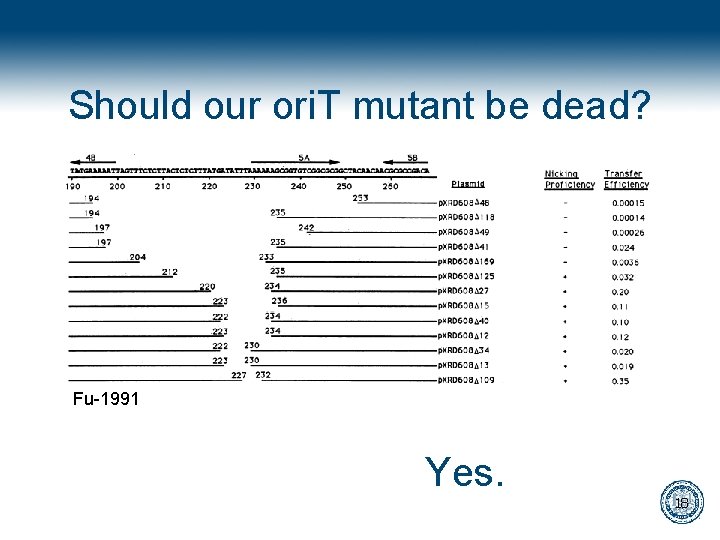
Should our ori. T mutant be dead? Fu-1991 Yes. 18
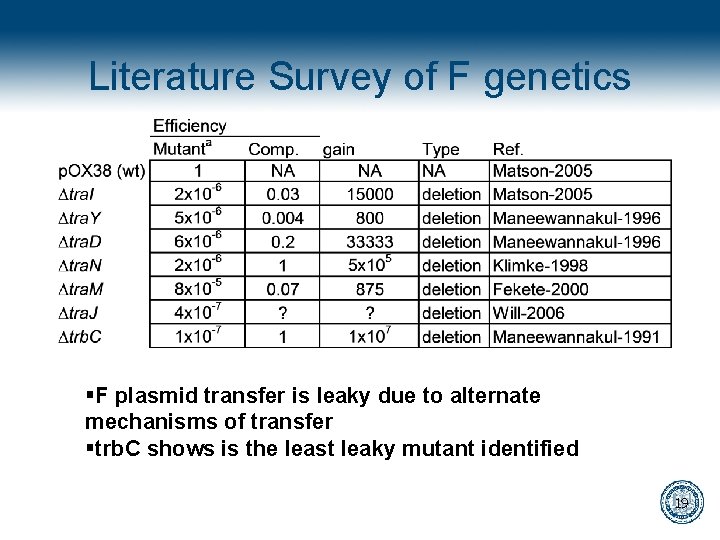
Literature Survey of F genetics §F plasmid transfer is leaky due to alternate mechanisms of transfer §trb. C shows is the least leaky mutant identified 19

Riboregulator Team 20
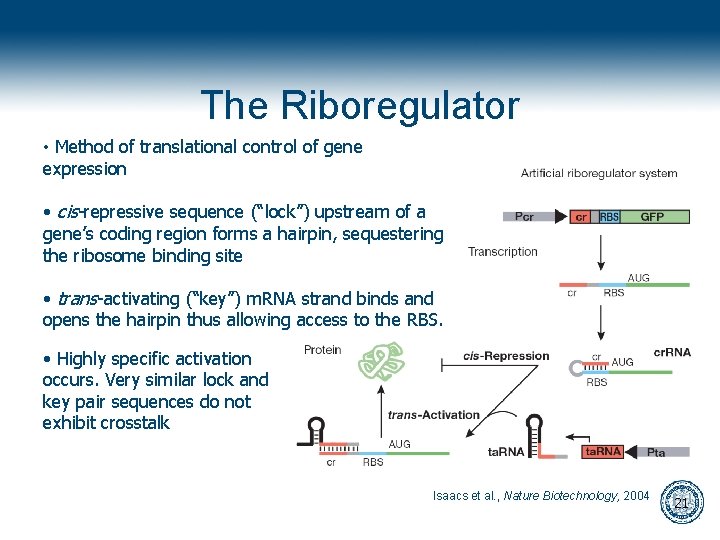
The Riboregulator • Method of translational control of gene expression • cis-repressive sequence (“lock”) upstream of a gene’s coding region forms a hairpin, sequestering the ribosome binding site • trans-activating (“key”) m. RNA strand binds and opens the hairpin thus allowing access to the RBS. • Highly specific activation occurs. Very similar lock and key pair sequences do not exhibit crosstalk Isaacs et al. , Nature Biotechnology, 2004 21
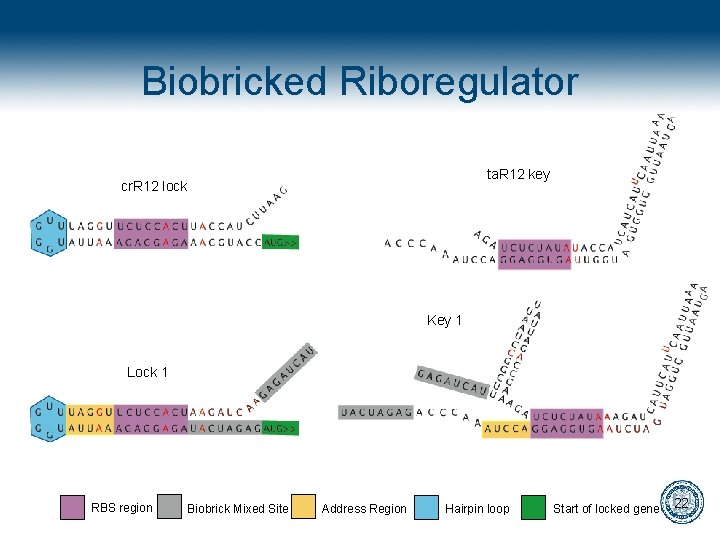
Biobricked Riboregulator ta. R 12 key cr. R 12 lock Key 1 Lock 1 RBS region Biobrick Mixed Site Address Region Hairpin loop Start of locked gene 22
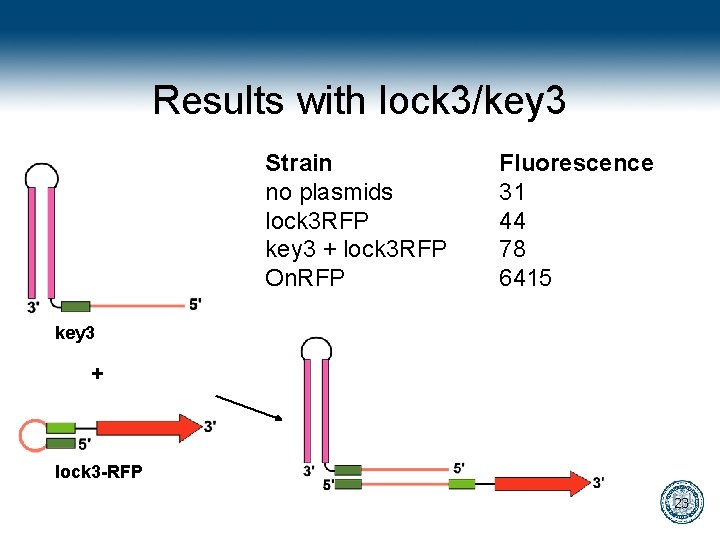
Results with lock 3/key 3 Strain no plasmids lock 3 RFP key 3 + lock 3 RFP On. RFP Fluorescence 31 44 78 6415 key 3 + lock 3 -RFP 23
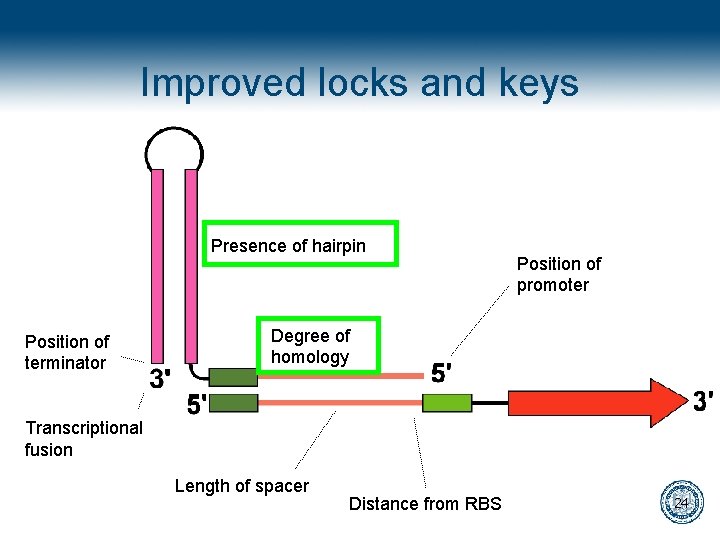
Improved locks and keys Presence of hairpin Position of terminator Position of promoter Degree of homology Transcriptional fusion Length of spacer Distance from RBS 24
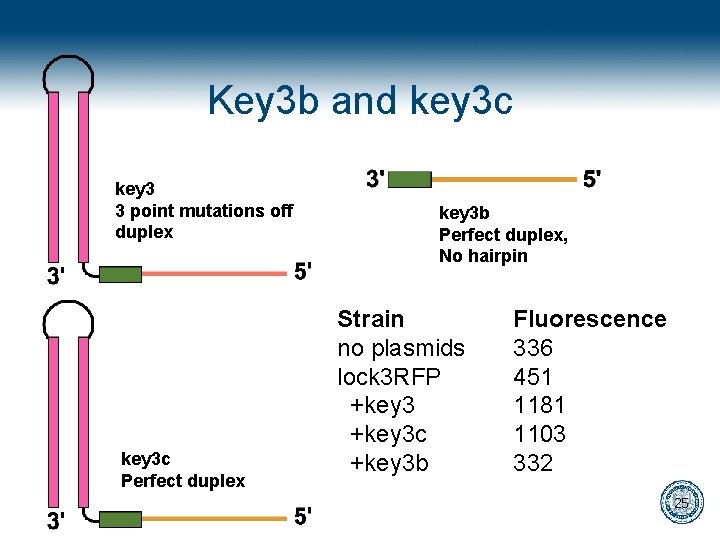
Key 3 b and key 3 c key 3 3 point mutations off duplex key 3 c Perfect duplex key 3 b Perfect duplex, No hairpin Strain no plasmids lock 3 RFP +key 3 c +key 3 b Fluorescence 336 451 1181 1103 332 25
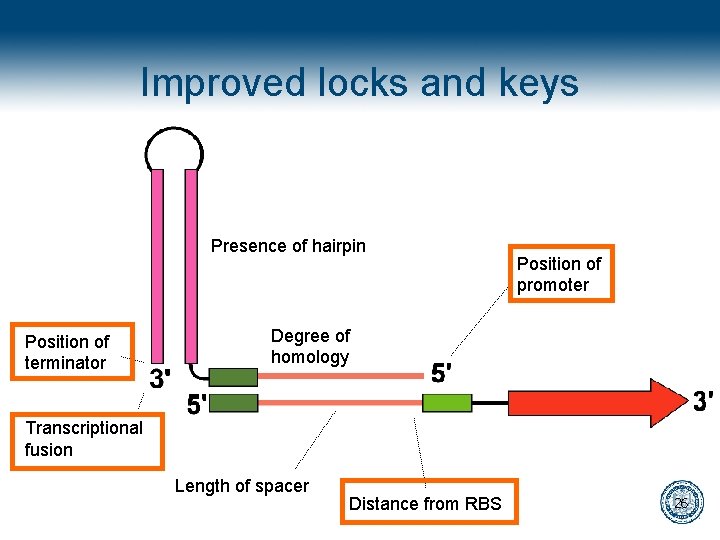
Improved locks and keys Presence of hairpin Position of terminator Position of promoter Degree of homology Transcriptional fusion Length of spacer Distance from RBS 26
Alternate hairpin structures key 3 d 27
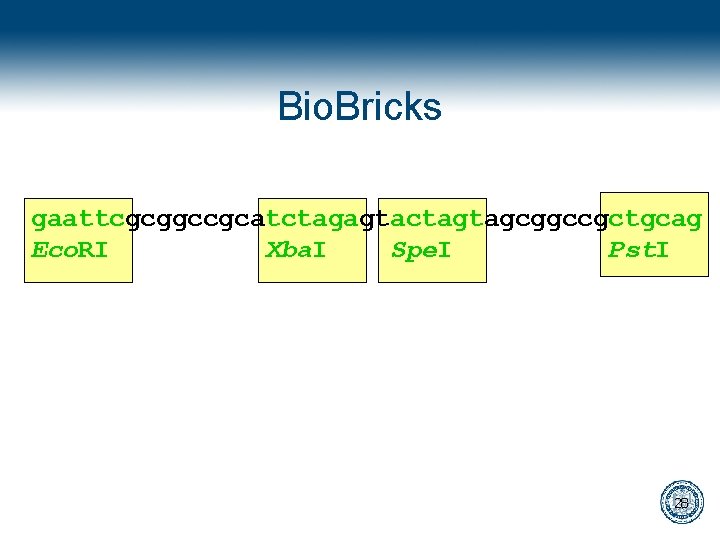
Bio. Bricks gaattcgcggccgcatctagagtactagtagcggccgctgcag Eco. RI Xba. I Spe. I Pst. I 28

gaattcgcggccgcatctagagtactagtagcggccgctgcag cttaagcgccggcgtagatctcatgatcatcgccggcgacgtc Digest gaattcgcggccgcat cttaagcgccggcgtagatc ctagtagcggccgctgcag atcgccggcgacgtc Ligate gaattcgcggccgcatctagtagcggccgctgcag cttaagcgccggcgtagatcatcgccggcgacgtc 29
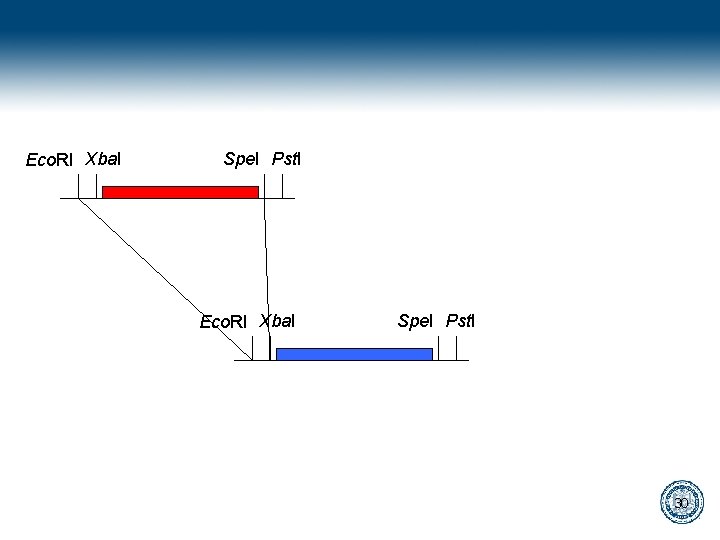
Eco. RI Xba. I Spe. I Pst. I 30
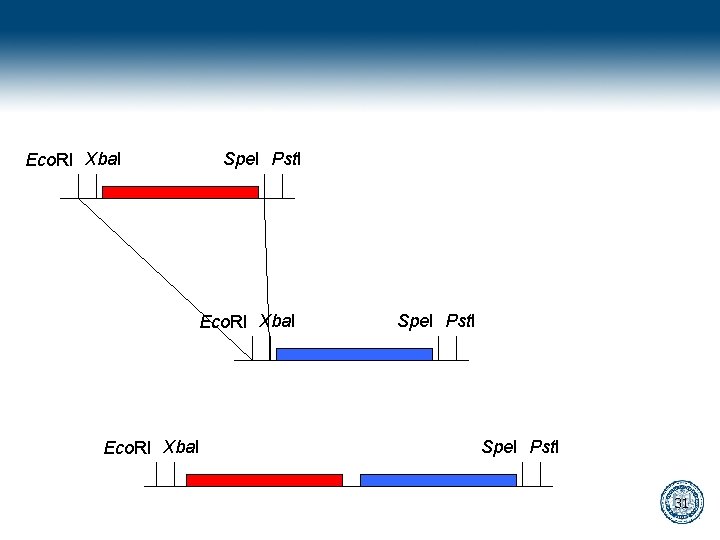
Eco. RI Xba. I Spe. I Pst. I 31
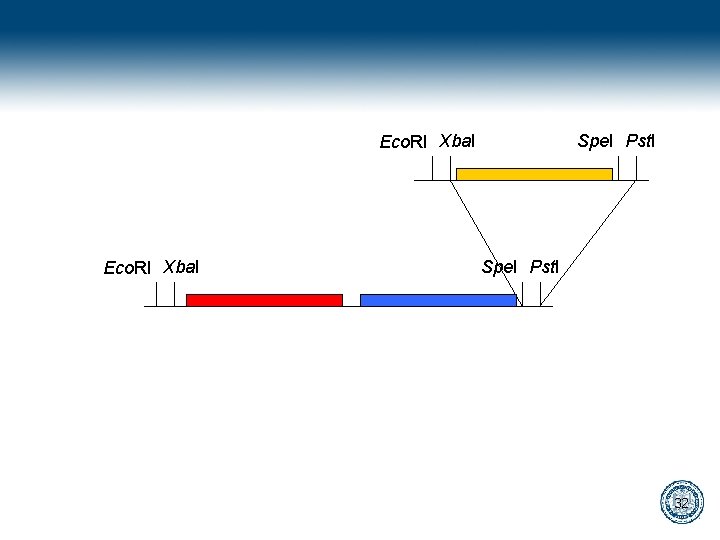
Eco. RI Xba. I Spe. I Pst. I 32
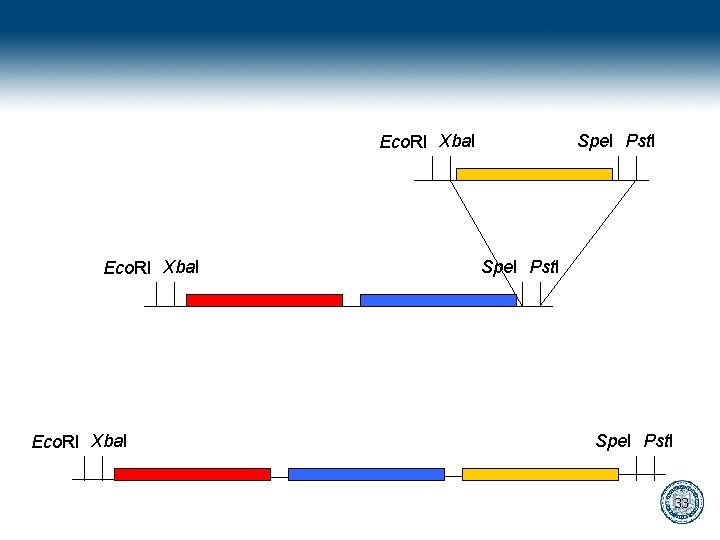
Eco. RI Xba. I Spe. I Pst. I 33
Biobrick plasmids: other origins p 15 A/Cm. R Biobrick p. SB 3 C 6 34
Functional suffixes and prefixes E-Ptet-X-SP p. J 23006 E-Ptet-rbs-X-SP EX-S-rbs. RFP-P 35
Suffix and prefix stuffers p. SB 1 A 2 -b 0015 p. SB 1 A? ? -b 0015 36

NAND Team 37
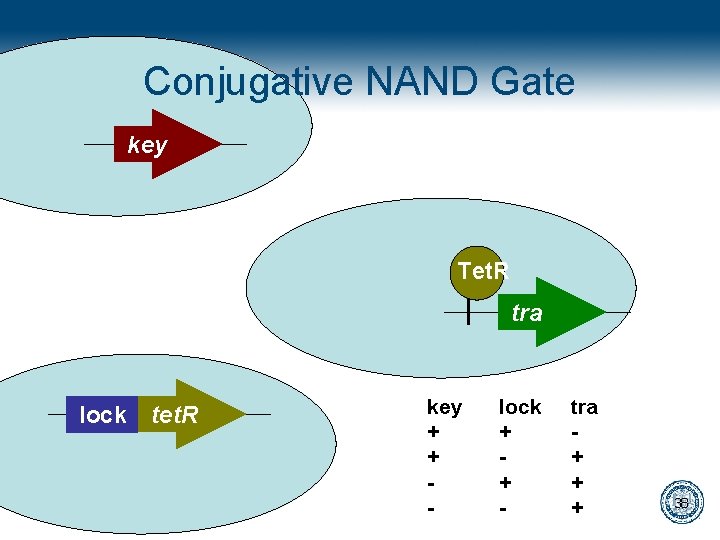
Conjugative NAND Gate key Tet. R tra lock tet. R key + + - lock + + - tra + + + 38
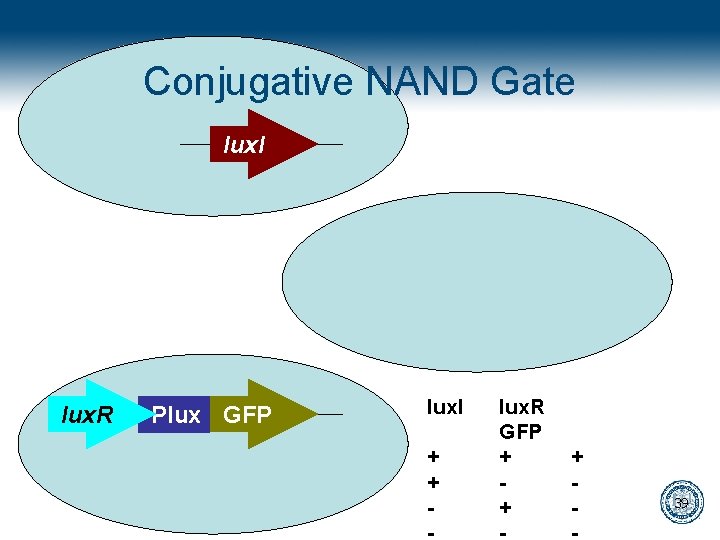
Conjugative NAND Gate key lux. I lux. R lock GFP Plux tet. R lux. I + + - lux. R GFP + + + - 39
The Wiki http: //www. openwetware. org/wiki/IGEM: UC_Berkeley/2006 40

Acknowledgements i. GEM-2005 team Jonathan Goler MIT folks: Randy Rettberg Reshma Shetty Melissa Li Keasling Lab Arkin Lab Microsoft for funding 41
- Slides: 41