Accessing U S Government Chemical Structure Databases with
Accessing U. S. Government Chemical Structure Databases with the CACTVS Toolkit Wolf-D. Ihlenfeldt Xemistry Gmb. H Lahntal, Germany wdi@xemistry. com
The US Gov Chemical Structure Data Information Pool o Pub. Chem n n Depositor structures (SID) Unique structures (CID) Assays (AID) Links to the rest of NCBI Entrez

The US Government Chemical Structure Data Pool o NIST Web Book n Spectra, physical properties o Chem. IDPlus n Phyical properties, biomedical links o NCI Chemical Identifier Resolver n From name and IDs to structure

Other sources o o o o o Chem. Spider (UK) EINECS (EU) KEGG (JP) EMolecules (US, commercial) Ch. EBI (UK) Ch. EMBL (UK) Drugbank (CA) PDB (US, academic) Common. Chemistry (US, commercial) Wikipedia (World)
How to Work with the Data? o Web interface for humans n Hard to work with software o Many DBs provide external links n Prone to breaking, becoming outdated o Data available as batch download n Massive, difficult to manage o Lack of formal interface documentation or programmatic access n Pub. Chem, Entrez, NCI Resolver good guys
The CACTVS Toolkit o Generic chemistry toolkit o Manages objects like structures, reactions, tables o Extensible collection of properties, methods and I/O modules o Implicit automatic method chaining o Scripting language interface for RAD o Ships with access properties and modules for all these databases o Comprehensive solution for multi-DB projects
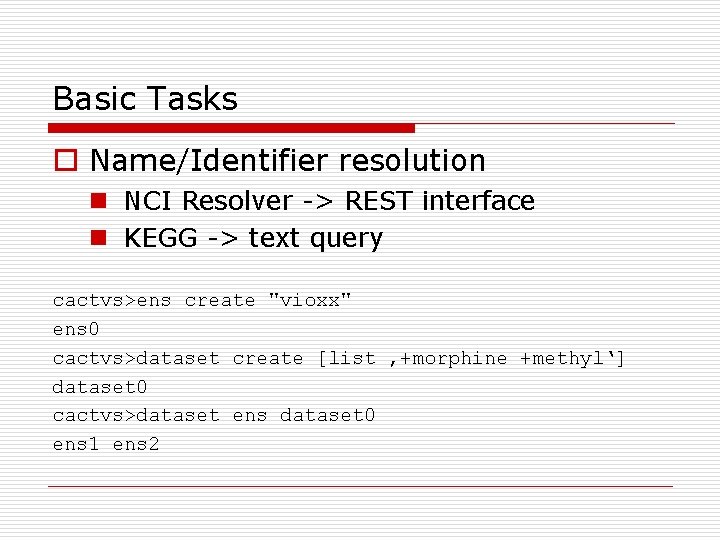
Basic Tasks o Name/Identifier resolution n NCI Resolver -> REST interface n KEGG -> text query cactvs>ens create "vioxx" ens 0 cactvs>dataset create [list ‚+morphine +methyl‘] dataset 0 cactvs>dataset ens dataset 0 ens 1 ens 2

Basic tasks: Get Database ID o Text structure query (SMILES, In. Ch. I) o NCBI PUG Web service cactvs>ens get ens 0 E_SIDSET 9792 207247 535364 5146347 7847634 7980536 8146414 8153131 10486532 11341940 11362123 11362973 11364757 11365535… cactvs>ens get ens 0 E_CHEMIDPLUS_ID 0162011907

Basic Tasks: Download Objects o Pub. Chem: from CID, SID, AID o PDB, CHEMBL, KEGG: from codes o Resolver: from name, identifiers cactvs>ens create 1 ens 0 cactvs>ens create CHEMBL 277500 ens 1

Basic Tasks: Download Objects o Pub. Chem I/O via native ASN. 1 cactvs>table create 198 table 3 cactvs>table get table 3 colnames SID_Source Version Date Outcome Score schedule endpoint vehicle dose tcprcnt toxicity cactvs> table get table 3 T_NCBI_ASSAY_DESCRIPTION(description) {The antitumor activity of compounds was measured in mice bearing transplantable tumors. Survival or tumor size were measured and the…
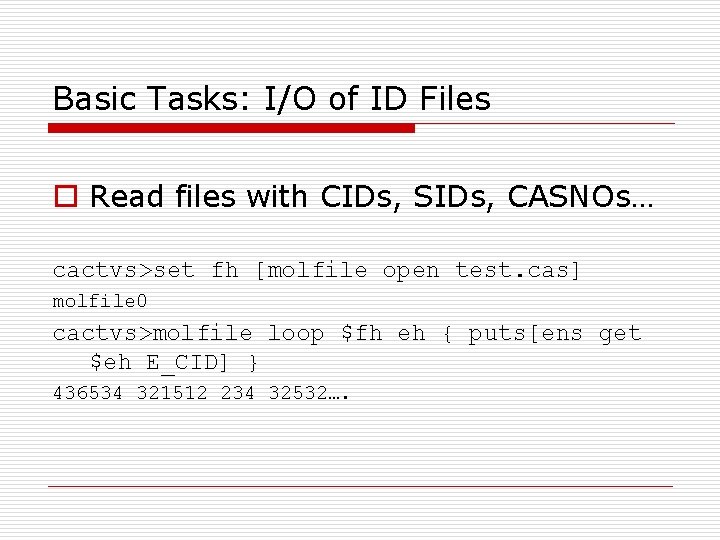
Basic Tasks: I/O of ID Files o Read files with CIDs, SIDs, CASNOs… cactvs>set fh [molfile open test. cas] molfile 0 cactvs>molfile loop $fh eh { puts[ens get $eh E_CID] } 436534 321512 234 32532….
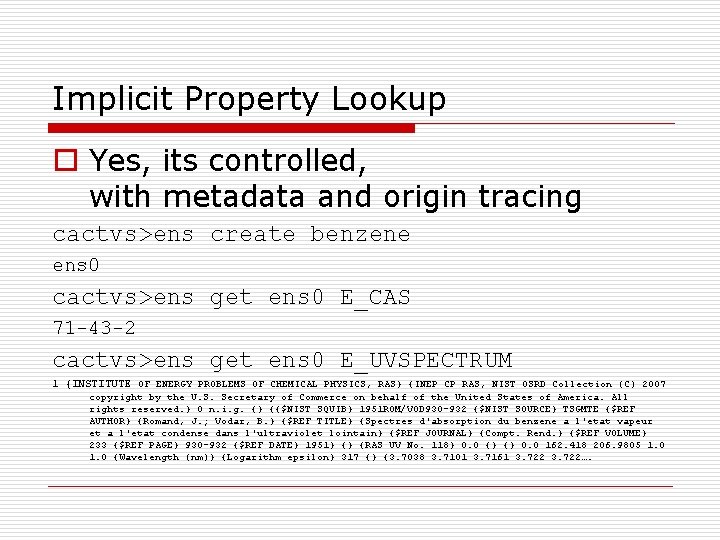
Implicit Property Lookup o Yes, its controlled, with metadata and origin tracing cactvs>ens create benzene ens 0 cactvs>ens get ens 0 E_CAS 71 -43 -2 cactvs>ens get ens 0 E_UVSPECTRUM 1 {INSTITUTE OF ENERGY PROBLEMS OF CHEMICAL PHYSICS, RAS} {INEP CP RAS, NIST OSRD Collection (C) 2007 copyright by the U. S. Secretary of Commerce on behalf of the United States of America. All rights reserved. } 0 n. i. g. {} {{$NIST SQUIB} 1951 ROM/VOD 930 -932 {$NIST SOURCE} TSGMTE {$REF AUTHOR} {Romand, J. ; Vodar, B. } {$REF TITLE} {Spectres d'absorption du benzene a l'etat vapeur et a l'etat condense dans l'ultraviolet lointain} {$REF JOURNAL} {Compt. Rend. } {$REF VOLUME} 233 {$REF PAGE} 930 -932 {$REF DATE} 1951} {} {RAS UV No. 118} 0. 0 {} {} 0. 0 162. 418 206. 9805 1. 0 {Wavelength (nm)} {Logarithm epsilon} 317 {} {3. 7038 3. 7101 3. 7161 3. 722….
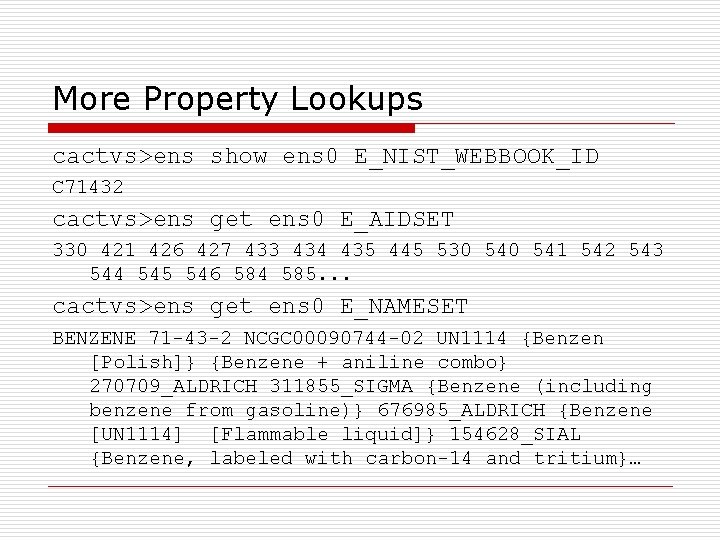
More Property Lookups cactvs>ens show ens 0 E_NIST_WEBBOOK_ID C 71432 cactvs>ens get ens 0 E_AIDSET 330 421 426 427 433 434 435 445 530 541 542 543 544 545 546 584 585. . . cactvs>ens get ens 0 E_NAMESET BENZENE 71 -43 -2 NCGC 00090744 -02 UN 1114 {Benzen [Polish]} {Benzene + aniline combo} 270709_ALDRICH 311855_SIGMA {Benzene (including benzene from gasoline)} 676985_ALDRICH {Benzene [UN 1114] [Flammable liquid]} 154628_SIAL {Benzene, labeled with carbon-14 and tritium}…
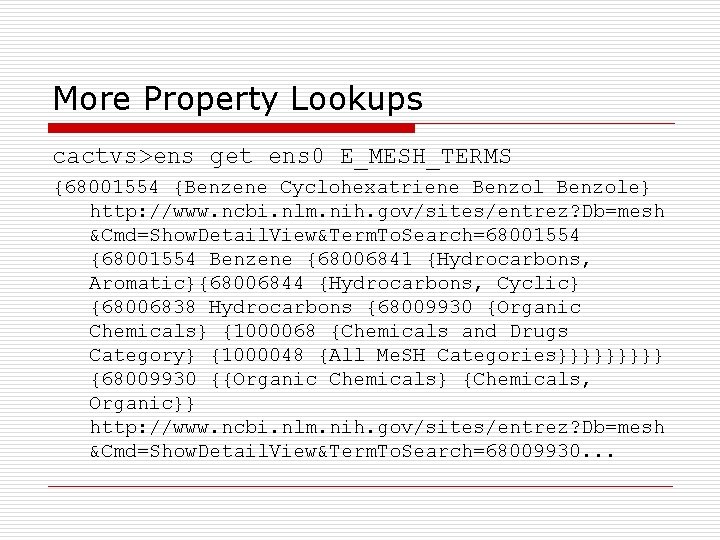
More Property Lookups cactvs>ens get ens 0 E_MESH_TERMS {68001554 {Benzene Cyclohexatriene Benzole} http: //www. ncbi. nlm. nih. gov/sites/entrez? Db=mesh &Cmd=Show. Detail. View&Term. To. Search=68001554 {68001554 Benzene {68006841 {Hydrocarbons, Aromatic}{68006844 {Hydrocarbons, Cyclic} {68006838 Hydrocarbons {68009930 {Organic Chemicals} {1000068 {Chemicals and Drugs Category} {1000048 {All Me. SH Categories}}}}} {68009930 {{Organic Chemicals} {Chemicals, Organic}} http: //www. ncbi. nlm. nih. gov/sites/entrez? Db=mesh &Cmd=Show. Detail. View&Term. To. Search=68009930. . .

Construction of Display URLs cactvs>ens get ens 0 E_PUBCHEM_URL http: //pubchem. ncbi. nlm. nih. gov/summary. cgi ? cid=241 cactvs>ens get ens 0 E_CHEMIDPLUS_URL http: //chem. sis. nlm. nih. gov/chemidplus/Proxy. Servlet ? object. Handle=DBMaint&action. Handle=default&next. P age=jsp/chemidheavy/Result. Screen. jsp&ROW_NUM=0&T XTSUPERLISTID=0000071432 cactvs>ens metadata ens 0 E_CHEMIDPLUS_URL info {JSESSIONID=257 C 452 AAB 26 D 395 DCC 4 AC 2652 F 05 C 99; Path=/chemidplus}

Ugliness under the Hood o Absence of a clean programmatic interface hurts! set mdata [encode -url [molfile string $eh]] set pdata "indexes=&DT_ROWS_PER_PAGE 2=1&object. Handle=Search&action. Handle=search. Chem. Id. Lite&next. Page=jsp%2 Fc hemidheavy%2 FChemid. Dataview. jsp&DT_ROWS_PER_PAGE=1&response. Handle=JSP&QV 10=&QO 10=Text+Search&QF 1 1=Locator&QV 11=&QO 11=in&STRING_TO_FILE=$mdata&QF 1=Name&QO 1=%3 D&QV 1=&QV 8=&QF 8=Tox. Test. Type&QO 5=bet ween&QV 5=&QF 5=Tox. Result&QV 6=&QF 6=Tox. Species&QV 7=&QF 7=Tox. Route&QV 9=&QF 9=Tox. Effect&Chem. Type=1001&Q F 3=Chem. Type&QV 3=&QF 2=Chem. Prop&QO 2=between&QV 2=&Chem. Data. Source. Type=0&QF 4=Chem. Data. Source. Type&QV 4=& Locator. Expr 1=&Locator. Oper=AND&Locator. Expr 2=&chemical_viewer=marvin&Structure. Similar. Pctg=80&QF 10= Structure. Equal&structure. Pref=marvin&QO 12=between&QV 12=&QF 12=Mol. Weight&x=22&y=5" set data [post -contenttype application/x-www-form-urlencoded -raw http: //chem. sis. nlm. nih. gov/chemidplus/Proxy. Servlet? chemidheavy $pdata #auto status] if {![regexp {chemid=([0 -9]+)} $data dummy id] && ![regexp {javascript: load. Chemical. Index[^0 -9]*([09]+)} $data dummy id]} { error "no Chem. IDplus record" } ens set $eh E_CHEMIDPLUS_ID $id ens metadata $eh E_CHEMIDPLUS_ID info $status(cookies)
Power by Design o In contrast, Pub. Chem has a welldefined set of interfaces – PUG, EUtils, cookie-free download URLs o No simulated Web form posting o No HTML page scraping o Support for more than just ID access
The Pub. Chem Virtual File Project o Improved access to Pub. Chem database indistinguishable from a local, read-only structure file in Cactvs scripting environment o Input functions transparently read structures and assay tables with all their data from Pub. Chem, by decoding native binary ASN. 1 o Query functions convenient development and conservation of queries exceeding the capabilites of Web interfaces and PUG, maintaining standard Cactvs query and retrieval syntax
Transforming the Pub. Chem Database into a Virtual File Cactvs toolkit uses file record as primary key Pub. Chem uses CID (AID, SID) as primary key Establish mapping via record/CID map Precomputed as 20 M bits bitmap Set bit indicates active CID Automatic download from Xemistry if needed, local caching, up-to-date check via Entrez query o Checked and potentially updated every 30 mins on Xemistry server o Data size 800 K compressed, download <10 s o Download of full active CID set from Entrez ~10 -25 mins o o o
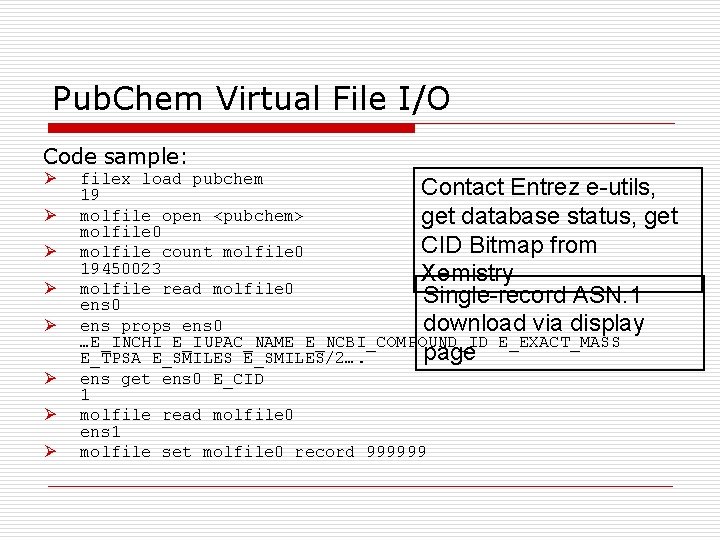
Pub. Chem Virtual File I/O Code sample: Ø Ø Ø Ø filex load pubchem Contact Entrez e-utils, 19 molfile open <pubchem> get database status, get molfile 0 CID Bitmap from molfile count molfile 0 19450023 Xemistry molfile read molfile 0 Single-record ASN. 1 ens 0 ens props ens 0 download via display …E_INCHI E_IUPAC_NAME E_NCBI_COMPOUND_ID E_EXACT_MASS page E_TPSA E_SMILES/2…. ens get ens 0 E_CID 1 molfile read molfile 0 ens 1 molfile set molfile 0 record 999999
![Simple Pub. Chem Queries Code sample: set fh [molfile open <pubchem>] set cidlist [molfile Simple Pub. Chem Queries Code sample: set fh [molfile open <pubchem>] set cidlist [molfile](http://slidetodoc.com/presentation_image_h2/b64657bff6d758c0d67fc2f86982b3ca/image-21.jpg)
Simple Pub. Chem Queries Code sample: set fh [molfile open <pubchem>] set cidlist [molfile scan $fh „structure >= $smarts“ {proplist E_CID}] Operations behind the scenes: o Set-up of PUG record o Post PUG, monitor return status o Cache CID result data o Direct access to result set, no structure download
![Intermediate Pub. Chem Queries Code sample: set fh [molfile open <pubchem>] set elist [molfile Intermediate Pub. Chem Queries Code sample: set fh [molfile open <pubchem>] set elist [molfile](http://slidetodoc.com/presentation_image_h2/b64657bff6d758c0d67fc2f86982b3ca/image-22.jpg)
Intermediate Pub. Chem Queries Code sample: set fh [molfile open <pubchem>] set elist [molfile scan $fh „or {structure = $smiles 1} {structure = $smiles 2} {structure = $smiles 3}“ enslist] Operations behind the scenes: o Create and post PUG records, get history keys o Perform server-side e-utils result merge via history keys o Retrieve CID set o Download structures as ASN. 1 blobs via CID
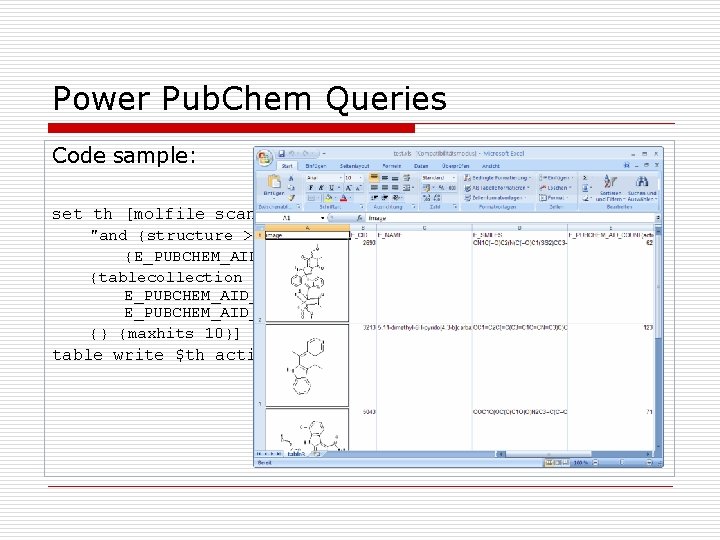
Power Pub. Chem Queries Code sample: set th [molfile scan <pubchem> "and {structure >= c 1 cncc 1} {E_PUBCHEM_AID_COUNT(active) > 25}„ {tablecollection image E_CID E_NAME E_SMILES E_PUBCHEM_AID_COUNT(active) E_PUBCHEM_AID_COUNT(inactive) E_ACTIVE_AIDSET} {} {maxhits 10}] table write $th active_pyrroles_in_pubchem. xls
Graphical Tools for the Masses o Draw or read structure o Compute database ID property n Display data o Compute lookup properties n Display data o Compute access URL property n Load page into HTML widget
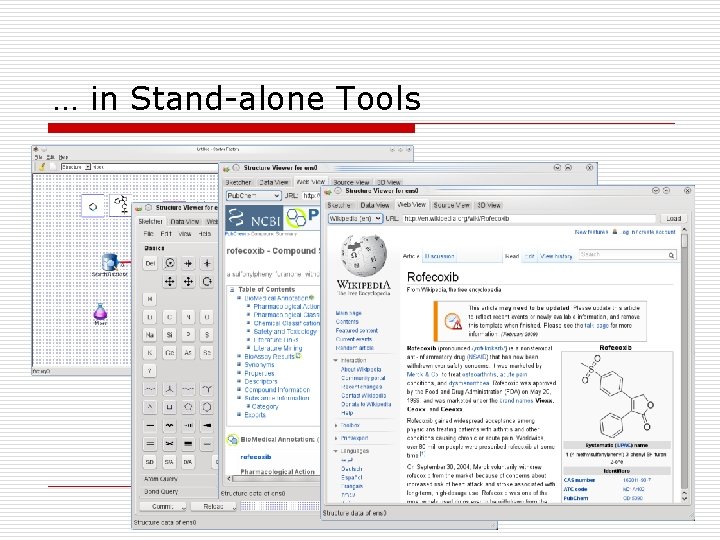
… in Stand-alone Tools
… and in Web Applications
- Slides: 26