Accessing Managing and Mining Unstructured Data Eugene Agichtein







![Page Quality: In Search of an Unbiased Web Ranking [Cho, Roy, Adams, SIGMOD 2005] Page Quality: In Search of an Unbiased Web Ranking [Cho, Roy, Adams, SIGMOD 2005]](https://slidetodoc.com/presentation_image_h2/ac8b084631e4d0edad06c5a8782564ff/image-33.jpg)


![Structure and evolution of blogspace [Kumar, Novak, Raghavan, Tomkins, CACM 2004, KDD 2006] Fraction Structure and evolution of blogspace [Kumar, Novak, Raghavan, Tomkins, CACM 2004, KDD 2006] Fraction](https://slidetodoc.com/presentation_image_h2/ac8b084631e4d0edad06c5a8782564ff/image-43.jpg)
![Structure of implicit entity-entity networks in text [Agichtein&Gravano, ICDE 2003] Connected Components Visualization Disease. Structure of implicit entity-entity networks in text [Agichtein&Gravano, ICDE 2003] Connected Components Visualization Disease.](https://slidetodoc.com/presentation_image_h2/ac8b084631e4d0edad06c5a8782564ff/image-44.jpg)
- Slides: 46

Accessing, Managing, and Mining Unstructured Data Eugene Agichtein 1

The Web n n n 20 B+ of machine-readable text (some of it useful) (Mostly) human-generated for human consumption q Both “artificial” and “natural” phenomenon Still growing? Local and global structure (links) Headaches: q Dynamic vs. static content q People figured out how to make money Positives: q Everything (almost) is on the web q People (eventually) can find info q People (on average) are not evil 2

Wait, there is more n n Blogs, wikipedia Hidden web: > 25 million databases q q q n n Accessible via keyword search interfaces E. g. , Med. Line, Cancer. Lit, USPTO, … 100 x more data than surface web (Transcribed) speech from Classified Genetic sequence annotations Biological & Medical literature Medical records, reports, alerts, 911 calls 3

Outline n Unstructured data (text, web, …) is q q Important (really!) Not so unstructured n Main tasks/requirements and challenges n Example problem: query optimization for text-centric tasks n Fundamental research problems/directions 4

Unstructured data = natural language text (for this talk) n Incredibly powerful and flexible means of communicating knowledge q n Local structures: syntax q q n Papers, news, web pages, lecture notes, patient records, shopping lists… English syntax HTML layout Semantics implicit, ambiguous, subjective I saw a man with a chainsaw n Need incredibly powerful and flexible decoder 5

Some more structure n Explicit link structure q Web, Blogs, Wikipedia, citations n Implicit link structure q Co-occurrence of entities within same document/context implies link between entities q Occurrence of same entity in multiple documents implies link between documents n Physical location q Page primarily “about” Atlanta q User somewhere around N. Decatur Rd q E-mail sender is two floors down n More on this later 6

Global Problem Space n n n n n Crawling (accessing) the data Storing (multiple version of) data “Understanding” the data information Indexing information Integration from multiple sources User-driven information retrieval Exploiting unstructured data in applications System-driven knowledge discovery Building a nuclear/hydro/wind/ power plant 7

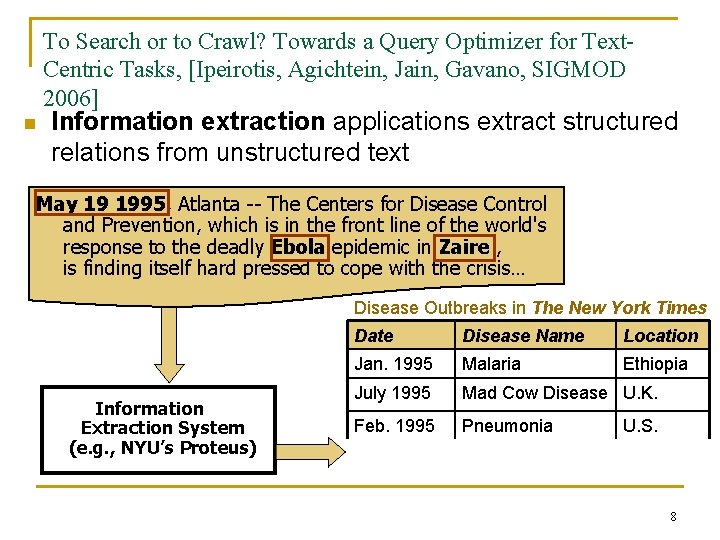
To Search or to Crawl? Towards a Query Optimizer for Text. Centric Tasks, [Ipeirotis, Agichtein, Jain, Gavano, SIGMOD 2006] n Information extraction applications extract structured relations from unstructured text May 19 1995, Atlanta -- The Centers for Disease Control and Prevention, which is in the front line of the world's response to the deadly Ebola epidemic in Zaire , is finding itself hard pressed to cope with the crisis… Disease Outbreaks in The New York Times Information Extraction System (e. g. , NYU’s Proteus) Date Disease Name Location Jan. 1995 Malaria Ethiopia July 1995 Mad Cow Disease U. K. Feb. 1995 Pneumonia U. S. May 1995 Ebola Zaire 8

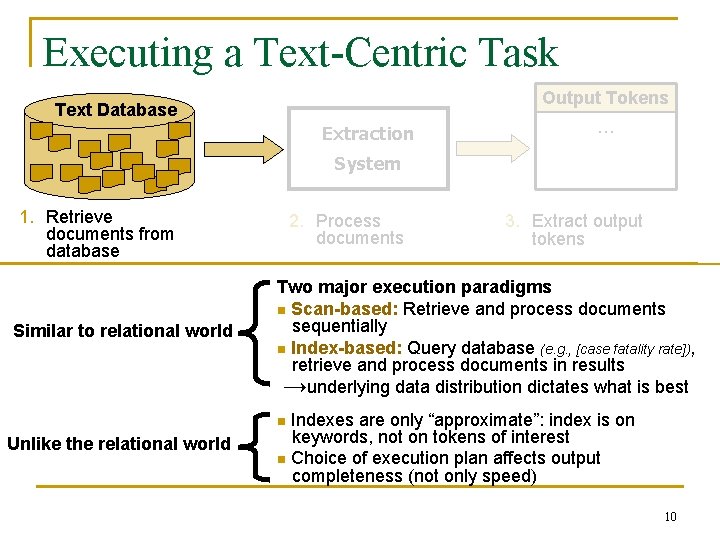
An Abstract View of Text-Centric Tasks Output Tokens Text Database … Extraction System 1. Retrieve documents from database 2. Process documents 3. Extract output tokens Task Token Information Extraction Relation Tuple Database Selection Word (+Frequency) Focused Crawling Web Page about a Topic For the rest of the talk 9

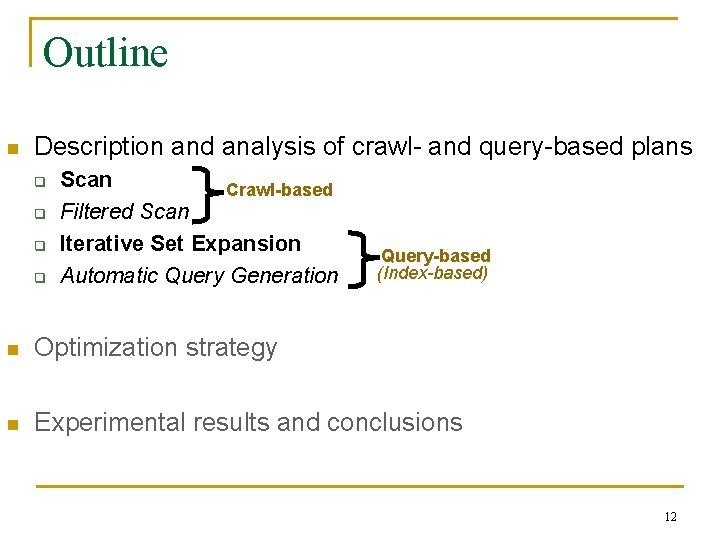
Executing a Text-Centric Task Output Tokens Text Database Extraction … System 1. Retrieve documents from database Similar to relational world 2. Process documents 3. Extract output tokens Two major execution paradigms n Scan-based: Retrieve and process documents sequentially n Index-based: Query database (e. g. , [case fatality rate]), retrieve and process documents in results →underlying data distribution dictates what is best Indexes are only “approximate”: index is on keywords, not on tokens of interest n Choice of execution plan affects output completeness (not only speed) n Unlike the relational world 10

Execution Plan Characteristics Output Tokens Text Database 1. Question: How do we choose the… Extraction fastest execution plan for reaching System a target recall ? Retrieve documents from database 2. Process documents 3. Extract output tokens Execution Plans have two main characteristics: n. Execution Time n. Recall (fraction of tokens retrieved) “What is the fastest plan for discovering 10% of the disease outbreaks mentioned in The New York Times archive? ” 11

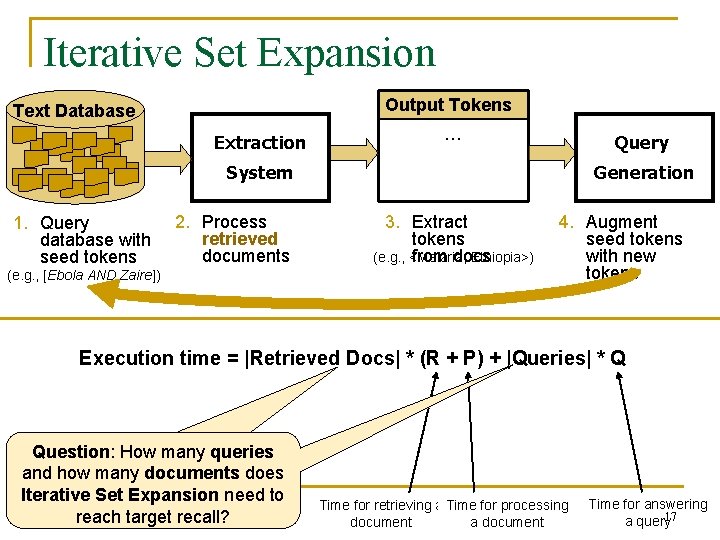
Outline n Description and analysis of crawl- and query-based plans q q Scan Crawl-based Filtered Scan Iterative Set Expansion Automatic Query Generation Query-based (Index-based) n Optimization strategy n Experimental results and conclusions 12

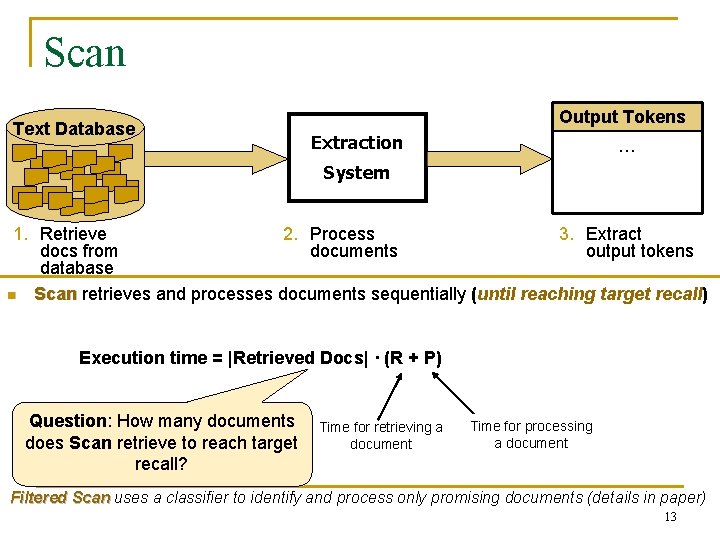
Scan Text Database Output Tokens Extraction … System 1. Retrieve 2. Process 3. Extract docs from documents output tokens database n Scan retrieves and processes documents sequentially (until reaching target recall) Execution time = |Retrieved Docs| · (R + P) Question: How many documents does Scan retrieve to reach target recall? Time for retrieving a document Time for processing a document Filtered Scan uses a classifier to identify and process only promising documents (details in paper) 13

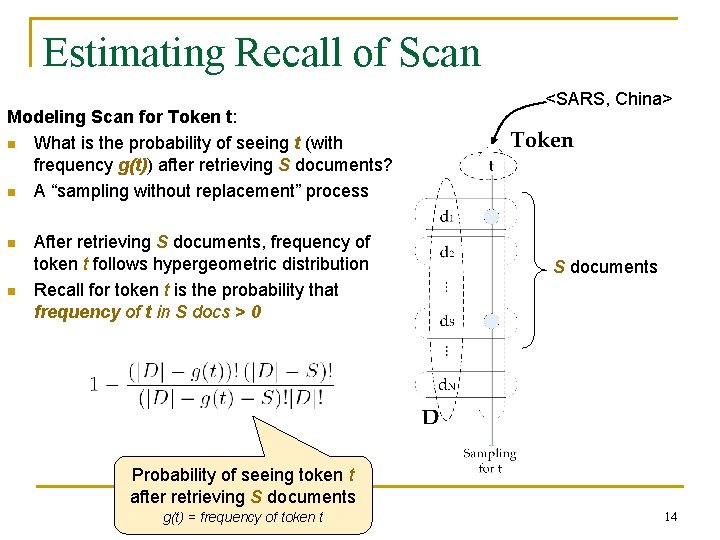
Estimating Recall of Scan Modeling Scan for Token t: n What is the probability of seeing t (with frequency g(t)) after retrieving S documents? n A “sampling without replacement” process n n After retrieving S documents, frequency of token t follows hypergeometric distribution Recall for token t is the probability that frequency of t in S docs > 0 <SARS, China> S documents Probability of seeing token t after retrieving S documents g(t) = frequency of token t 14

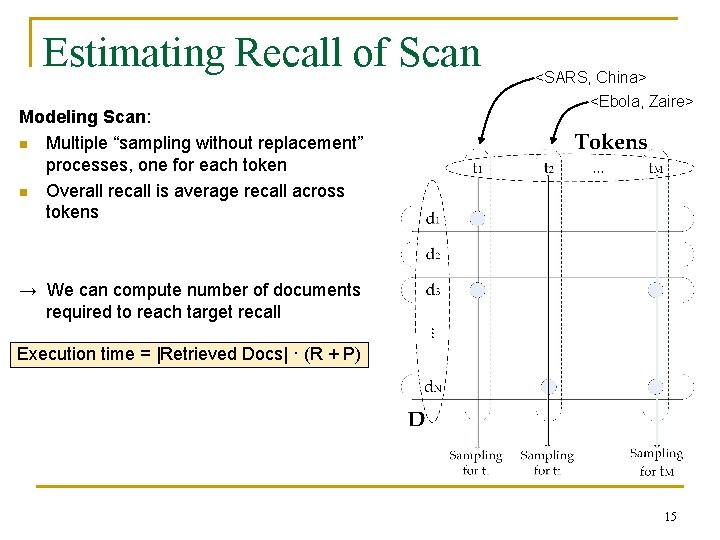
Estimating Recall of Scan Modeling Scan: n Multiple “sampling without replacement” processes, one for each token n Overall recall is average recall across tokens <SARS, China> <Ebola, Zaire> → We can compute number of documents required to reach target recall Execution time = |Retrieved Docs| · (R + P) 15

Outline n Description and analysis of crawl- and query-based plans q q Scan Crawl-based Filtered Scan Iterative Set Expansion Automatic Query Generation Query-based n Optimization strategy n Experimental results and conclusions 16

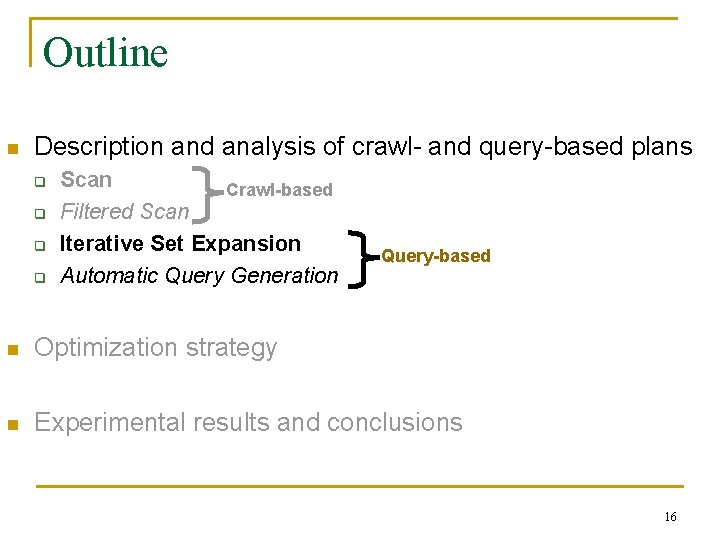
Iterative Set Expansion Output Tokens Text Database Extraction … Query System 1. Query database with seed tokens 2. Process retrieved documents (e. g. , [Ebola AND Zaire]) Generation 3. Extract tokens (e. g. , <Malaria, Ethiopia>) from docs 4. Augment seed tokens with new tokens Execution time = |Retrieved Docs| * (R + P) + |Queries| * Q Question: How many queries and how many documents does Iterative Set Expansion need to reach target recall? Time for retrieving a Time for processing document a document Time for answering a query 17

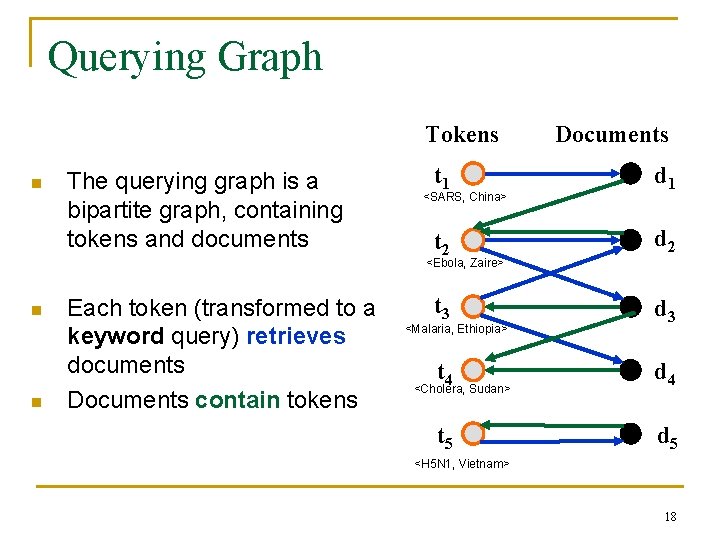
Querying Graph Tokens n The querying graph is a bipartite graph, containing tokens and documents Documents t 1 d 1 t 2 d 2 t 3 d 3 <SARS, China> <Ebola, Zaire> n n Each token (transformed to a keyword query) retrieves documents Documents contain tokens <Malaria, Ethiopia> t 4 d 4 t 5 d 5 <Cholera, Sudan> <H 5 N 1, Vietnam> 18

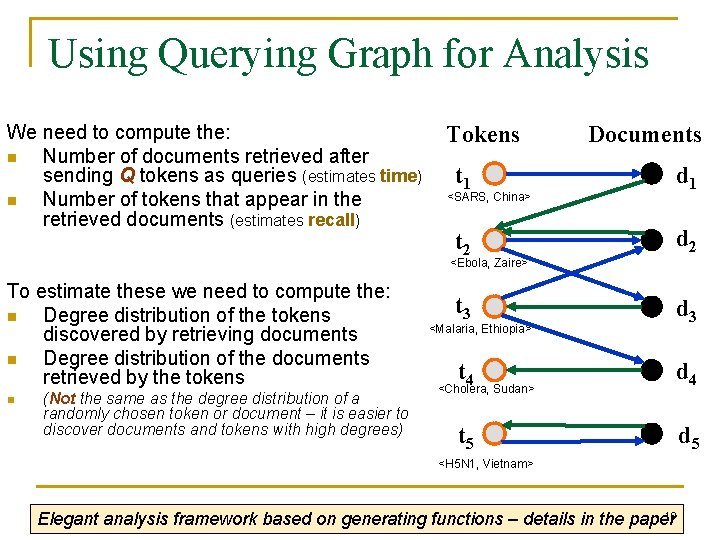
Using Querying Graph for Analysis We need to compute the: n Number of documents retrieved after sending Q tokens as queries (estimates time) n Number of tokens that appear in the retrieved documents (estimates recall) Tokens Documents t 1 d 1 t 2 d 2 t 3 d 3 <SARS, China> <Ebola, Zaire> To estimate these we need to compute the: n Degree distribution of the tokens discovered by retrieving documents n Degree distribution of the documents retrieved by the tokens n (Not the same as the degree distribution of a randomly chosen token or document – it is easier to discover documents and tokens with high degrees) <Malaria, Ethiopia> t 4 d 4 t 5 d 5 <Cholera, Sudan> <H 5 N 1, Vietnam> 19 Elegant analysis framework based on generating functions – details in the paper

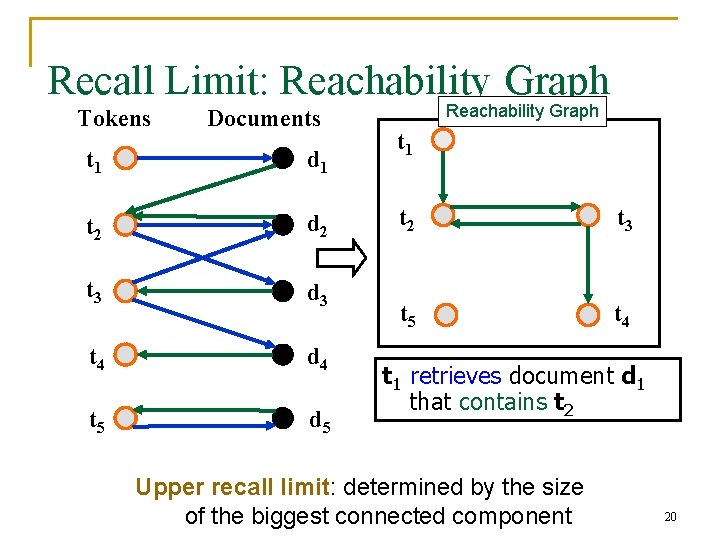
Recall Limit: Reachability Graph Tokens Documents t 1 d 1 t 2 d 2 t 3 d 3 t 4 d 4 t 5 d 5 Reachability Graph t 1 t 2 t 3 t 5 t 4 t 1 retrieves document d 1 that contains t 2 Upper recall limit: determined by the size of the biggest connected component 20

Automatic Query Generation n Iterative Set Expansion has recall limitation due to iterative nature of query generation n Automatic Query Generation avoids this problem by creating queries offline (using machine learning), which are designed to return documents with tokens Details in the papers 21

Outline n Description and analysis of crawl- and query-based plans n Optimization strategy n Experimental results and conclusions 22

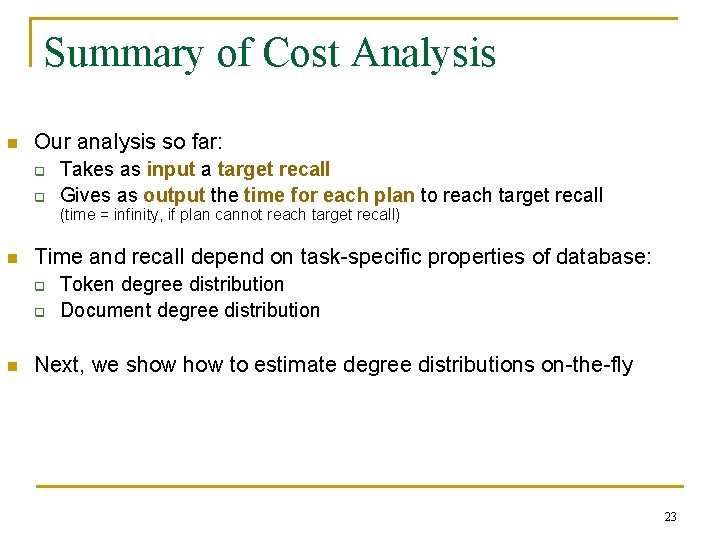
Summary of Cost Analysis n Our analysis so far: q q Takes as input a target recall Gives as output the time for each plan to reach target recall (time = infinity, if plan cannot reach target recall) n Time and recall depend on task-specific properties of database: q q n Token degree distribution Document degree distribution Next, we show to estimate degree distributions on-the-fly 23

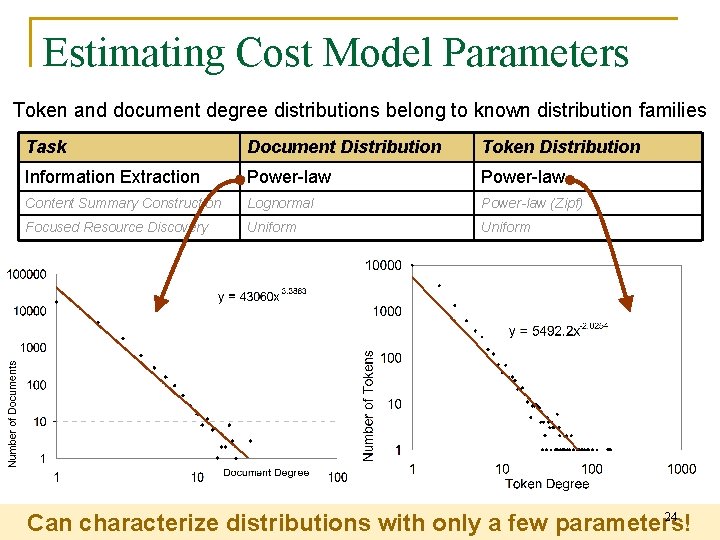
Estimating Cost Model Parameters Token and document degree distributions belong to known distribution families Task Document Distribution Token Distribution Information Extraction Power-law Content Summary Construction Lognormal Power-law (Zipf) Focused Resource Discovery Uniform 24 Can characterize distributions with only a few parameters!

Parameter Estimation n Naïve solution for parameter estimation: q q q Start with separate, “parameter-estimation” phase Perform random sampling on database Stop when cross-validation indicates high confidence n We can do better than this! n No need for separate sampling phase Sampling is equivalent to executing the task: →Piggyback parameter estimation into execution n 25

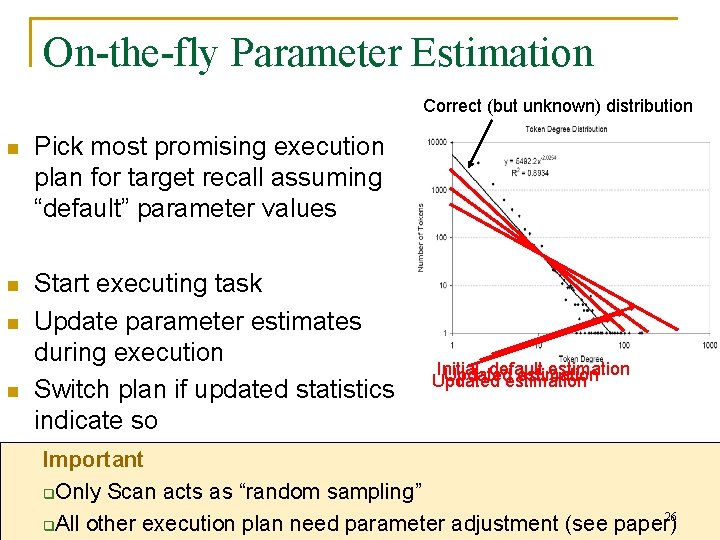
On-the-fly Parameter Estimation Correct (but unknown) distribution n Pick most promising execution plan for target recall assuming “default” parameter values n Start executing task Update parameter estimates during execution Switch plan if updated statistics indicate so n n Initial, default estimation Updated estimation Important q. Only Scan acts as “random sampling” 26 q. All other execution plan need parameter adjustment (see paper)

Outline n Description and analysis of crawl- and query-based plans n Optimization strategy n Experimental results and conclusions 27

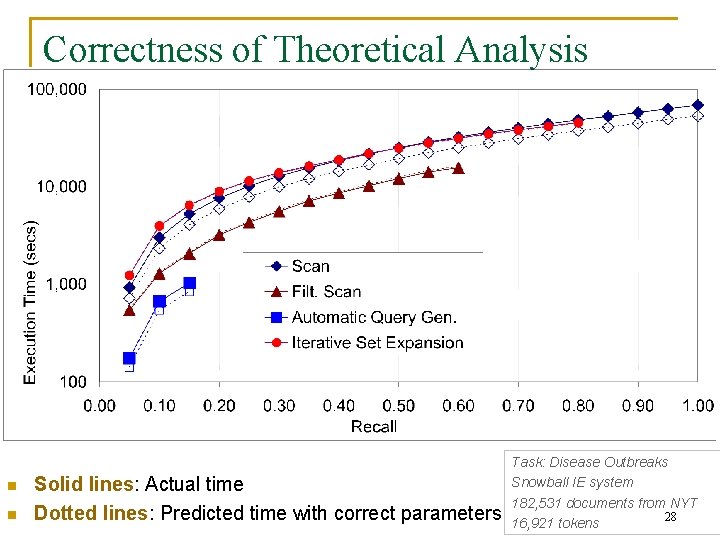
Correctness of Theoretical Analysis n n Solid lines: Actual time Dotted lines: Predicted time with correct parameters Task: Disease Outbreaks Snowball IE system 182, 531 documents from NYT 28 16, 921 tokens

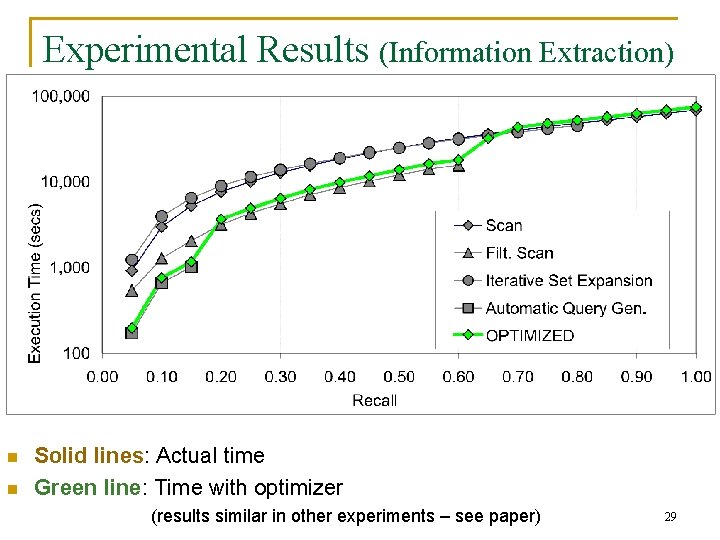
Experimental Results (Information Extraction) n n Solid lines: Actual time Green line: Time with optimizer (results similar in other experiments – see paper) 29

Conclusions n Common execution plans for multiple text-centric tasks n Analytic models for predicting execution time and recall of various crawl- and query-based plans n Techniques for on-the-fly parameter estimation n Optimization framework picks on-the-fly the fastest plan for target recall 30

Global Problem Space n n n n Crawling (accessing) the data “Understand” the data information Indexing information Integration from multiple sources User-driven information retrieval Exploiting unstructured data in applications System-driven knowledge discovery 31

Some Research Directions n Modeling explicit and Implicit network structures q q q n Knowledge Discovery from Biological and Medical Data q q n Automatic sequence annotation bioinformatics, genetics Actionable knowledge extraction from medical articles Robust information extraction, retrieval, and query processing q q q n Modeling evolution of explicit structure on web, blogspace, wikipedia Modeling implicit link structures in text, collections, web Exploiting implicit & explicit social networks (e. g. , for epidemiology) Integrating information in structured and unstructured sources Robust search/question answering for medical applications Confidence estimation for extraction from text and other sources Detecting reliable signals from (noisy) text data (e. g. , : medical surveillance) Accuracy (!=authority) of online sources Information diffusion/propagation in online sources q q Information propagation on the web In collaborative sources (wikipedia, Med. Line) 32
![Page Quality In Search of an Unbiased Web Ranking Cho Roy Adams SIGMOD 2005 Page Quality: In Search of an Unbiased Web Ranking [Cho, Roy, Adams, SIGMOD 2005]](https://slidetodoc.com/presentation_image_h2/ac8b084631e4d0edad06c5a8782564ff/image-33.jpg)
Page Quality: In Search of an Unbiased Web Ranking [Cho, Roy, Adams, SIGMOD 2005] n “popular pages tend to get even more popular, while unpopular pages get ignored by an average user” 33

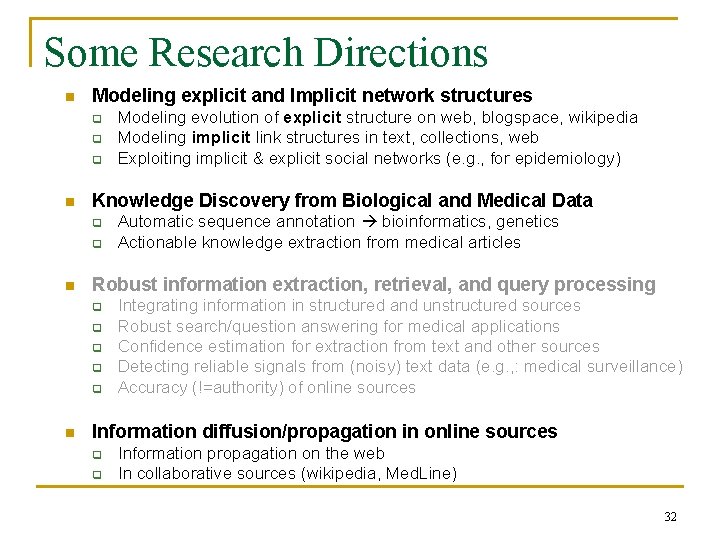
Sic Transit Gloria Telae: Towards an Understanding of the Web’s Decay [Bar-Yossef, Broder, Kumar, Tomkins, WWW 2004] 34

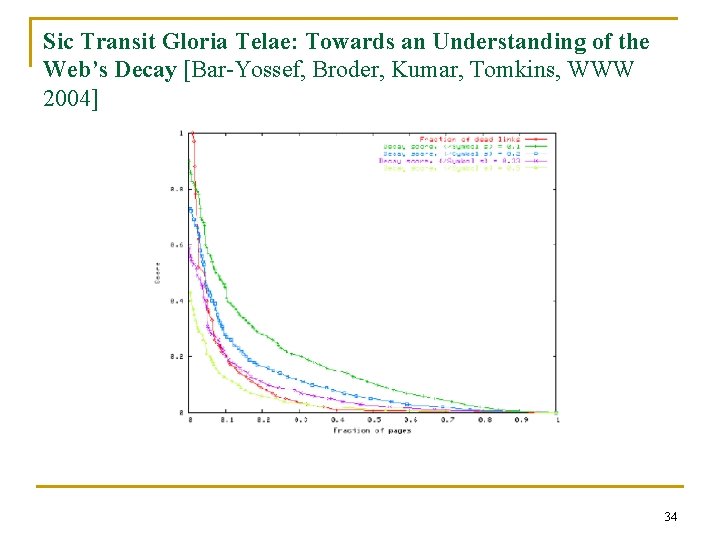
Modeling Social Networks for n Epidemiology, security, … Email exchange mapped onto cubicle locations. 35

Some Research Directions n Modeling explicit and Implicit network structures q q q n Knowledge Discovery from Biological and Medical Data q q n Automatic sequence annotation bioinformatics, genetics Actionable knowledge extraction from medical articles Robust information extraction, retrieval, and query processing q q q n Modeling evolution of explicit structure on web, blogspace, wikipedia Modeling implicit link structures in text, collections, web Exploiting implicit & explicit social networks (e. g. , for epidemiology) Integrating information in structured and unstructured sources Query processing over unstructured text Robust search/question answering for medical applications Confidence estimation for extraction from text and other sources Detecting reliable signals from (noisy) text data (e. g. , : medical surveillance) Information diffusion/propagation in online sources q q Information propagation on the web In collaborative sources (wikipedia, Med. Line) 36

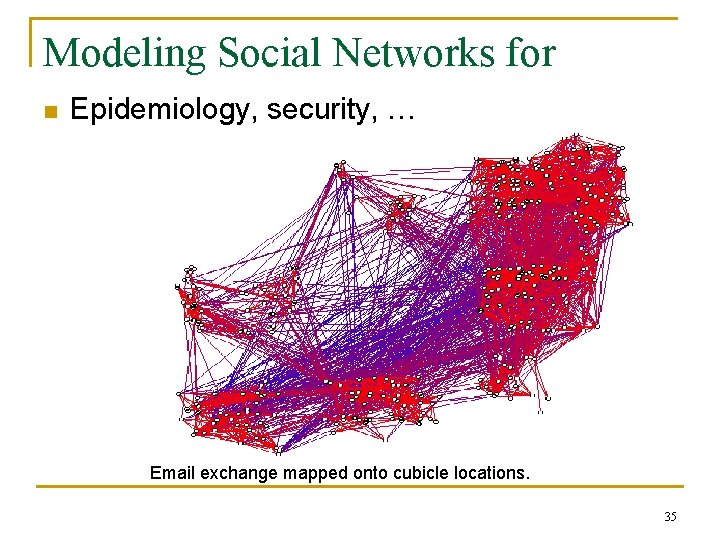
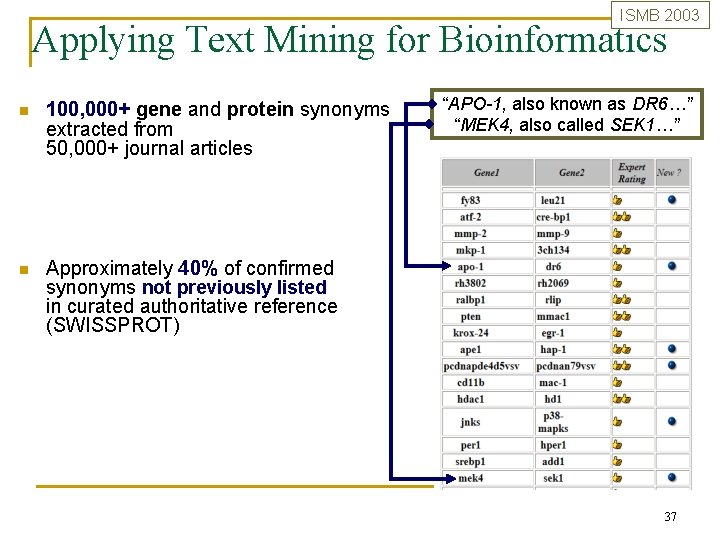
ISMB 2003 Applying Text Mining for Bioinformatics n 100, 000+ gene and protein synonyms extracted from 50, 000+ journal articles n Approximately 40% of confirmed synonyms not previously listed in curated authoritative reference (SWISSPROT) “APO-1, also known as DR 6…” “MEK 4, also called SEK 1…” 37

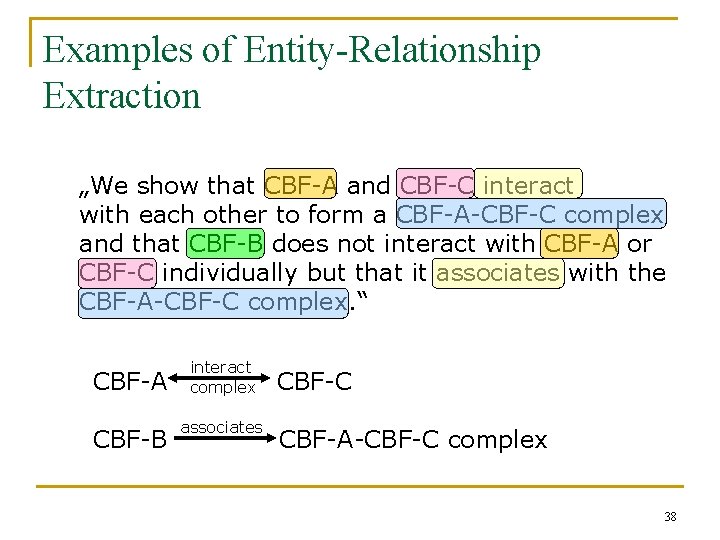
Examples of Entity-Relationship Extraction „We show that CBF-A and CBF-C interact with each other to form a CBF-A-CBF-C complex and that CBF-B does not interact with CBF-A or CBF-C individually but that it associates with the CBF-A-CBF-C complex. “ CBF-A CBF-B interact complex associates CBF-C CBF-A-CBF-C complex 38

Another Example Z-100 is an arabinomannan extracted from Mycobacterium tuberculosis that has various immunomodulatory activities, such as the induction of interleukin 12, interferon gamma (IFN-gamma) and beta-chemokines. The effects of Z-100 on human immunodeficiency virus type 1 (HIV-1) replication in human monocyte-derived macrophages (MDMs) are investigated in this paper. In MDMs, Z-100 markedly suppressed the replication of not only macrophage-tropic (M-tropic) HIV-1 strain (HIV-1 JR-CSF), but also HIV-1 pseudotypes that possessed amphotropic Moloney murine leukemia virus or vesicular stomatitis virus G envelopes. Z-100 was found to inhibit HIV-1 expression, even when added 24 h after infection. In addition, it substantially inhibited the expression of the p. NL 43 luc. Deltaenv vector (in which the env gene is defective and the nef gene is replaced with the firefly luciferase gene) when this vector was transfected directly into MDMs. These findings suggest that Z-100 inhibits virus replication, mainly at HIV-1 transcription. However, Z 100 also downregulated expression of the cell surface receptors CD 4 and CCR 5 in MDMs, suggesting some inhibitory effect on HIV-1 entry. Further experiments revealed that Z-100 induced IFN-beta production in these cells, resulting in induction of the 16 -k. Da CCAAT/enhancer binding protein (C/EBP) beta transcription factor that represses HIV-1 long terminal repeat transcription. These effects were alleviated by SB 203580, a specific inhibitor of p 38 mitogen-activated protein kinases (MAPK), indicating that the p 38 MAPK signalling pathway was involved in Z-100 -induced repression of HIV-1 replication in MDMs. These findings suggest that Z-100 might be a useful immunomodulator for control of HIV-1 infection. 39

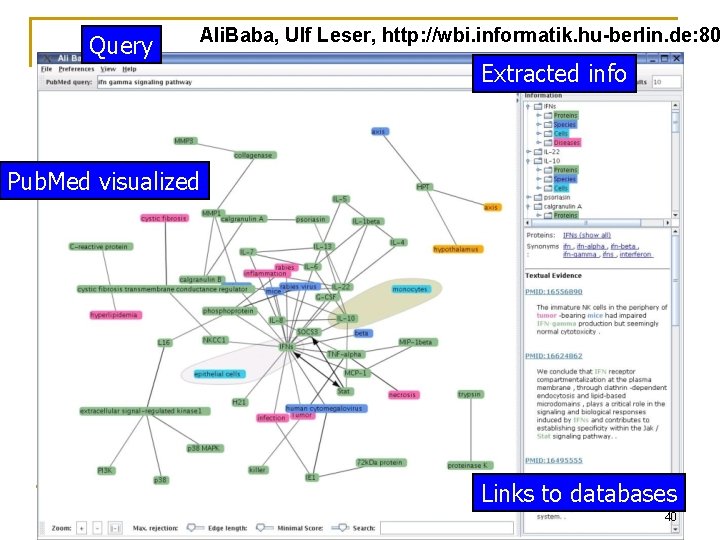
Query Ali. Baba, Ulf Leser, http: //wbi. informatik. hu-berlin. de: 80 Extracted info Pub. Med visualized Links to databases 40

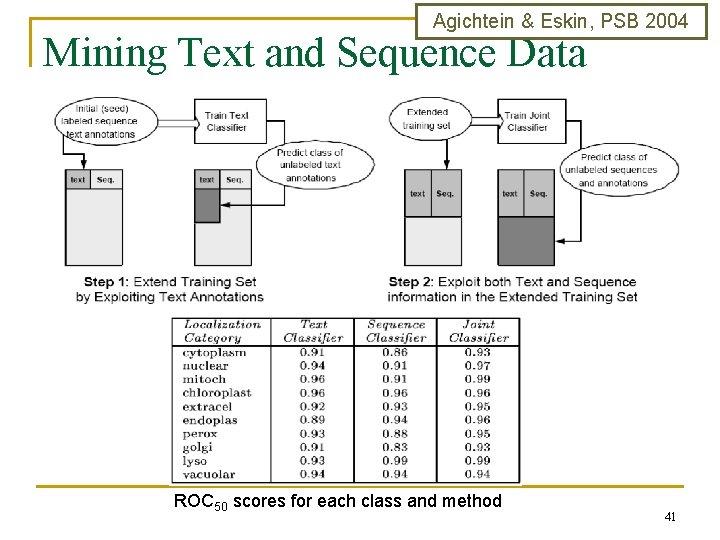
Agichtein & Eskin, PSB 2004 Mining Text and Sequence Data ROC 50 scores for each class and method 41

Some Research Directions n Modeling explicit and Implicit network structures q q q n Knowledge Discovery from Biological and Medical Data q q n Automatic sequence annotation bioinformatics, genetics Actionable knowledge extraction from medical articles Robust information extraction, retrieval, and query processing q q q n Modeling evolution of explicit structure on web, blogspace, wikipedia Modeling implicit link structures in text, collections, web Exploiting implicit & explicit social networks (e. g. , for epidemiology) Integrating information in structured and unstructured sources Robust search/question answering for medical applications Confidence estimation for extraction from text and other sources Detecting reliable signals from (noisy) text data (e. g. , : medical surveillance) Accuracy (!=authority) of online sources Information diffusion/propagation in online sources q q Information propagation on the web In collaborative sources (wikipedia, Med. Line) 42
![Structure and evolution of blogspace Kumar Novak Raghavan Tomkins CACM 2004 KDD 2006 Fraction Structure and evolution of blogspace [Kumar, Novak, Raghavan, Tomkins, CACM 2004, KDD 2006] Fraction](https://slidetodoc.com/presentation_image_h2/ac8b084631e4d0edad06c5a8782564ff/image-43.jpg)
Structure and evolution of blogspace [Kumar, Novak, Raghavan, Tomkins, CACM 2004, KDD 2006] Fraction of nodes in components of various sizes within Flickr and Yahoo! 360 timegraph, by week. 43
![Structure of implicit entityentity networks in text AgichteinGravano ICDE 2003 Connected Components Visualization Disease Structure of implicit entity-entity networks in text [Agichtein&Gravano, ICDE 2003] Connected Components Visualization Disease.](https://slidetodoc.com/presentation_image_h2/ac8b084631e4d0edad06c5a8782564ff/image-44.jpg)
Structure of implicit entity-entity networks in text [Agichtein&Gravano, ICDE 2003] Connected Components Visualization Disease. Outbreaks, 44 New York Times 1995

Some Research Directions n Modeling explicit and Implicit network structures q q q n Knowledge Discovery from Biological and Medical Data q q n Automatic sequence annotation bioinformatics, genetics Actionable knowledge extraction from medical articles Robust information extraction, retrieval, and query processing q q q n Modeling evolution of explicit structure on web, blogspace, wikipedia Modeling implicit link structures in text, collections, web Exploiting implicit & explicit social networks (e. g. , for epidemiology) Integrating information in structured and unstructured sources Robust search/question answering for medical applications Confidence estimation for extraction from text and other sources Detecting reliable signals from (noisy) text data (e. g. , : medical surveillance) Accuracy (!=authority) of online sources Information diffusion/propagation in online sources q q Information propagation on the web, news In collaborative sources (wikipedia, Med. Line) 45

Thank You n Details: http: //www. mathcs. emory. edu/~eugene/ 46