Access Reactome Database via Application Programming Interfaces APIs

Access Reactome Database via Application Programming Interfaces (APIs) Guanming Wu Ontario Institute for Cancer Research
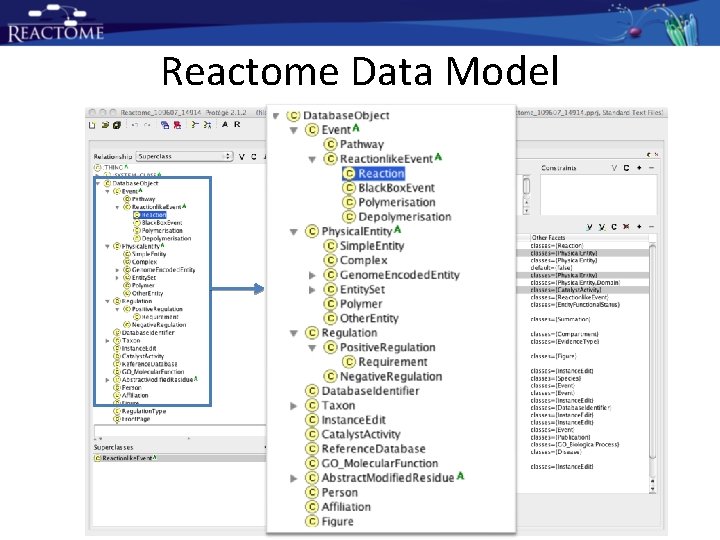
Reactome Data Model
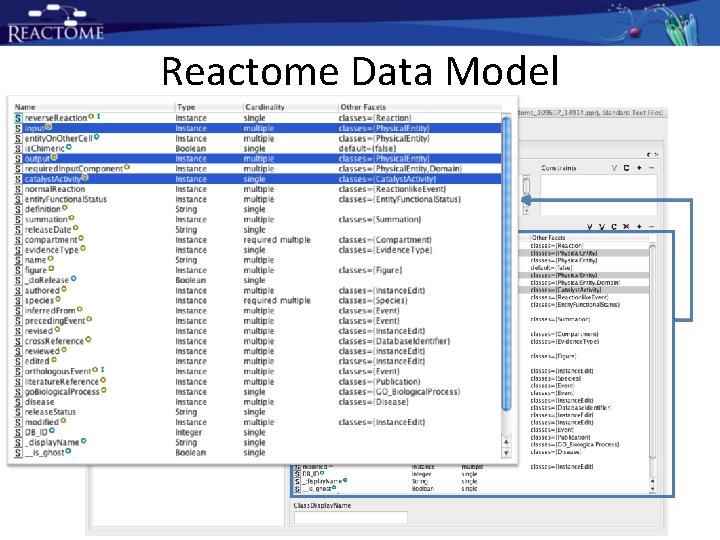
Reactome Data Model
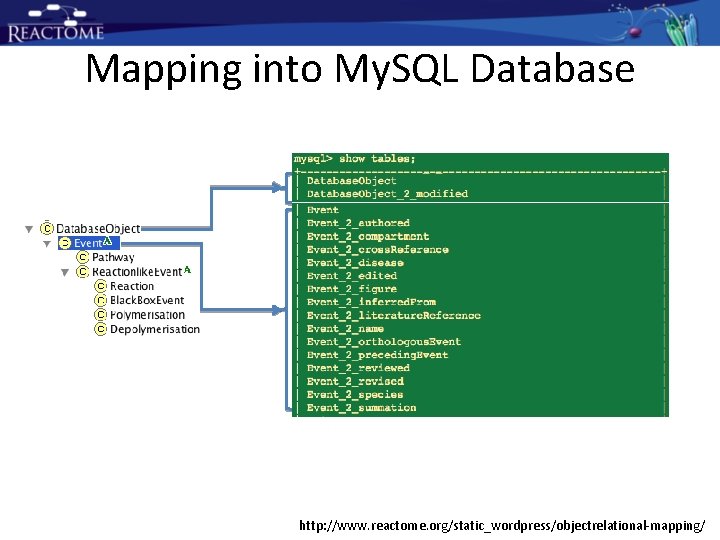
Mapping into My. SQL Database http: //www. reactome. org/static_wordpress/objectrelational-mapping/
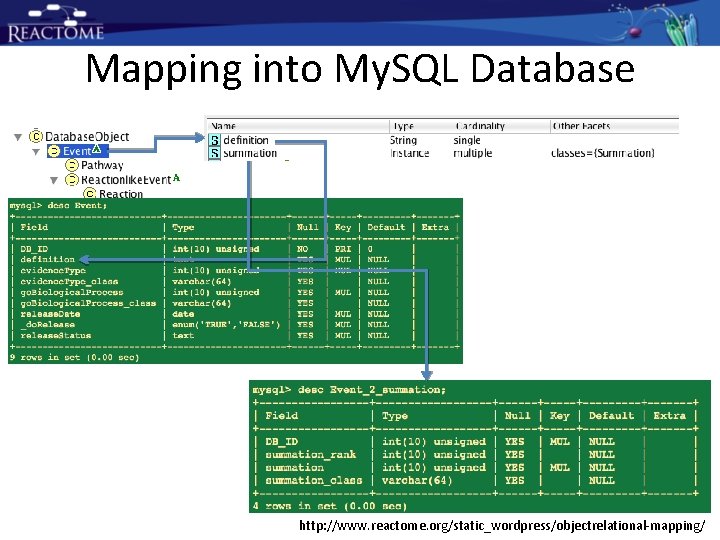
Mapping into My. SQL Database http: //www. reactome. org/static_wordpress/objectrelational-mapping/
Reactome APIs • • Java API Perl API SOAP API RESTful API
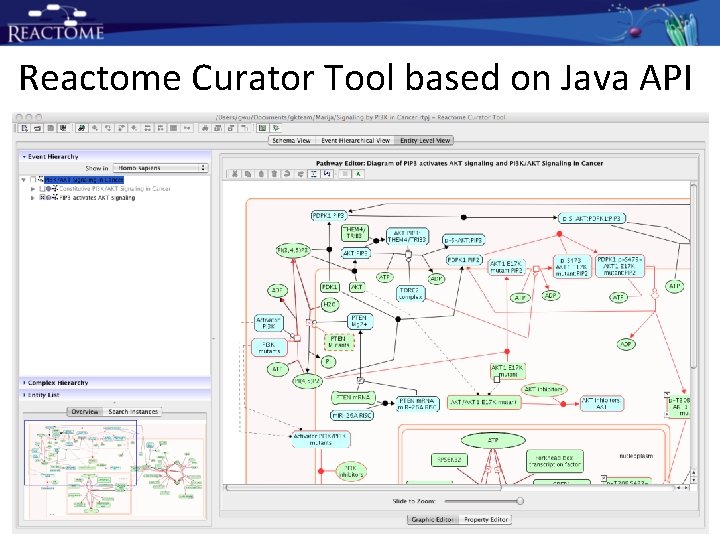
Reactome Curator Tool based on Java API

Get Reactome Data and Java API http: //www. reactome. org/download/index. html • Download a mysql database dump file and load into a mysql database – sql • Download the Reactome curator tool – reactome. jar: add it to your classpath – src. jar: Java source code for reactome. jar – mysql. jar: JDBC driver
Java Data Model: Schema

Java Data Model: Class

Java Data Model: Attribute
Java Data Model: Instance
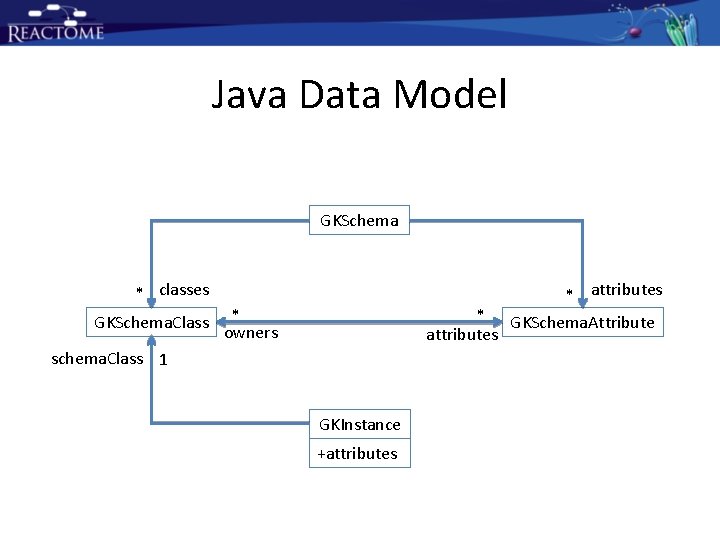
Java Data Model GKSchema * classes GKSchema. Class * owners * attributes schema. Class 1 GKInstance +attributes * attributes GKSchema. Attribute
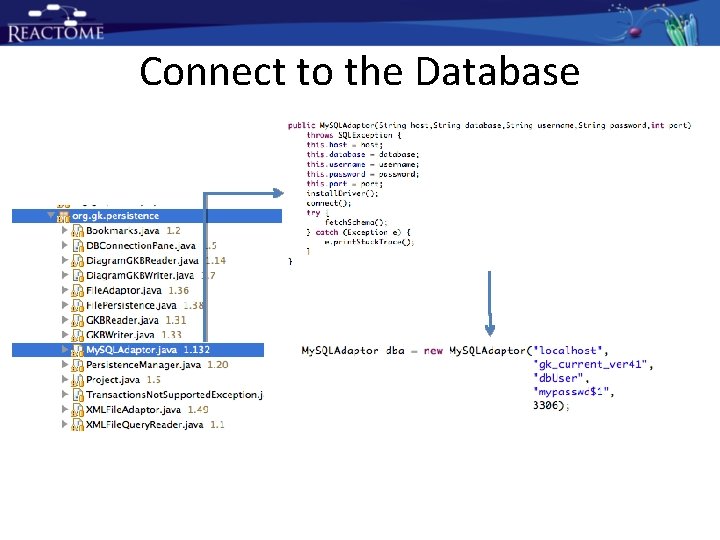
Connect to the Database

Use My. SQLAdaptor Class

Use My. SQLAdaptor Class
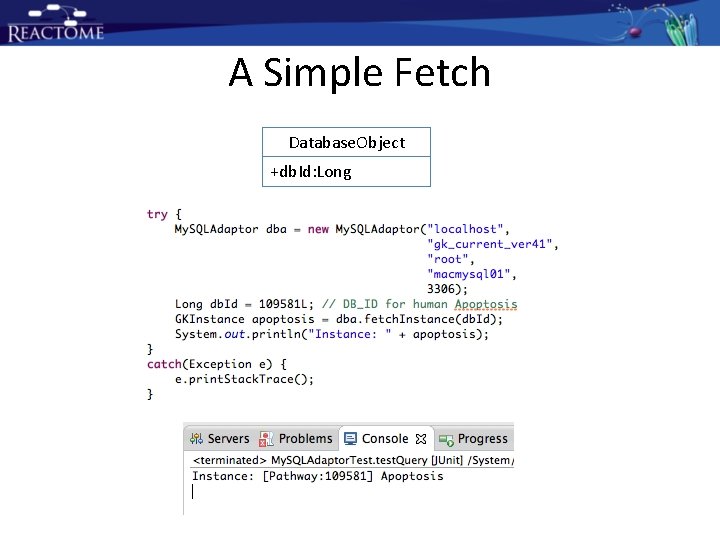
A Simple Fetch Database. Object +db. Id: Long
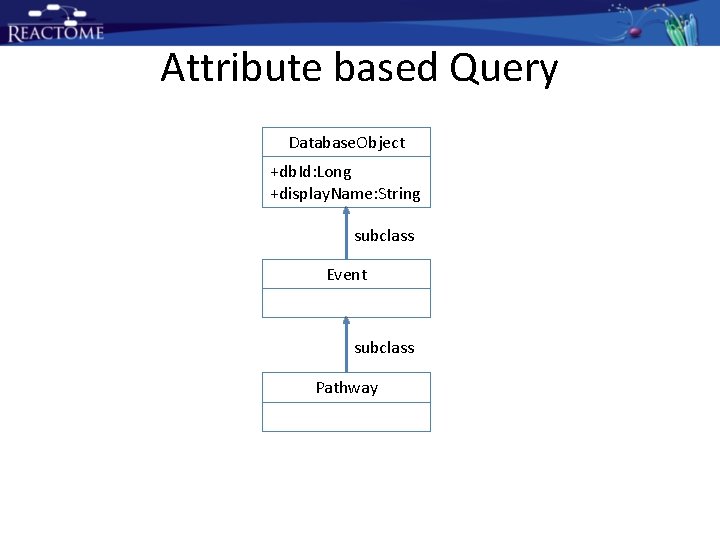
Attribute based Query Database. Object +db. Id: Long +display. Name: String subclass Event subclass Pathway

Attribute based Query Operator may be “like” or others supported by SQL or mysql

Attribute based Query
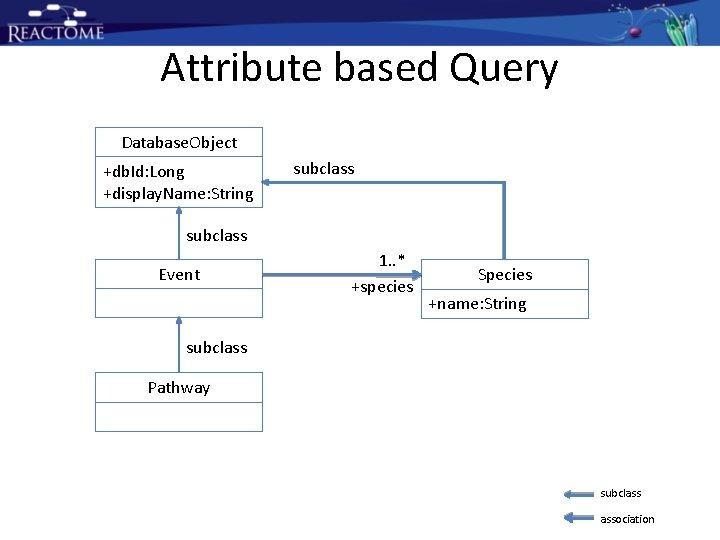
Attribute based Query Database. Object +db. Id: Long +display. Name: String subclass Event 1. . * +species Species +name: String subclass Pathway subclass association

Attribute based Query
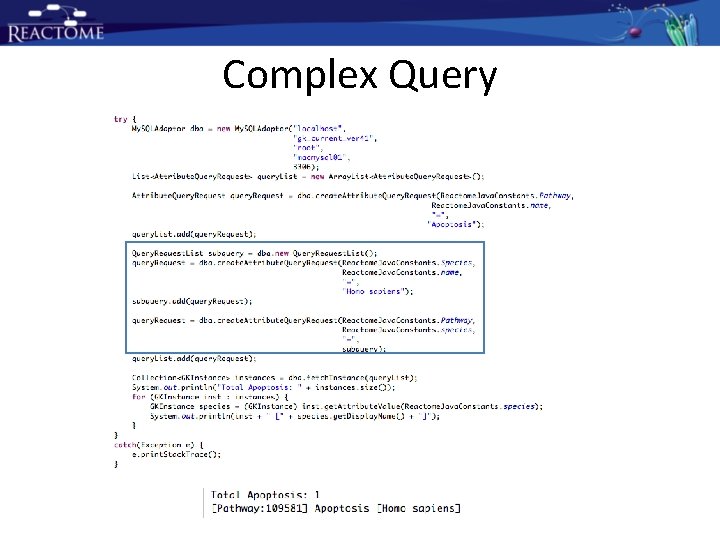
Complex Query
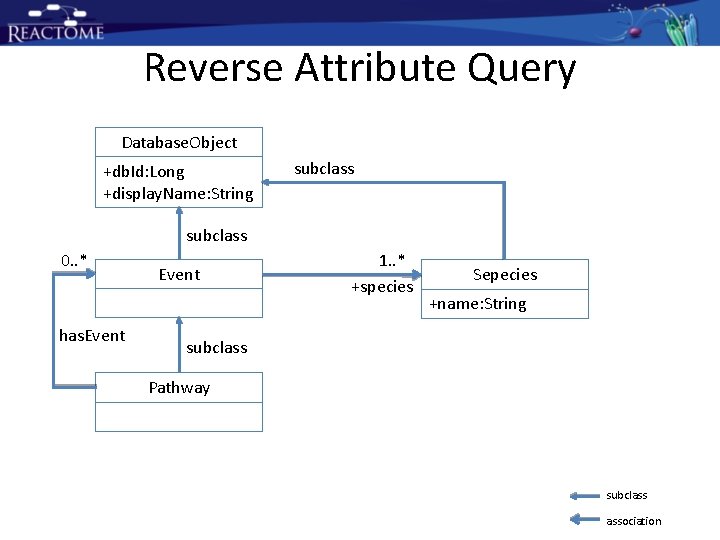
Reverse Attribute Query Database. Object +db. Id: Long +display. Name: String subclass 0. . * has. Event 1. . * +species Sepecies +name: String subclass Pathway subclass association
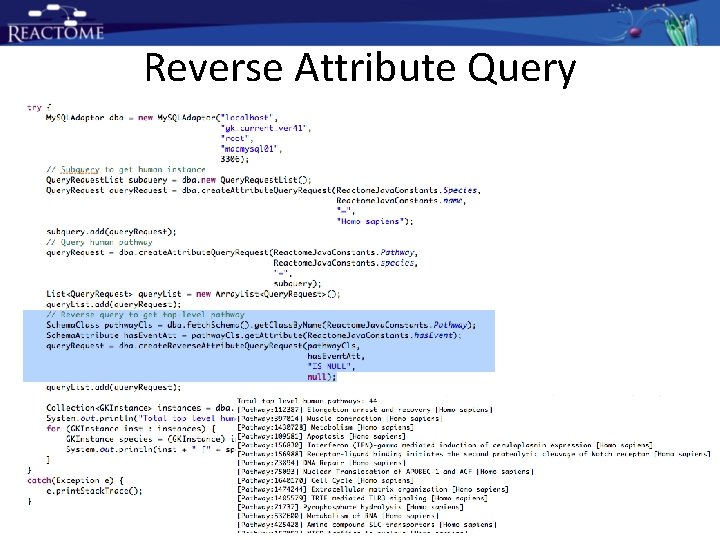
Reverse Attribute Query

Another Reverse Attribute Query
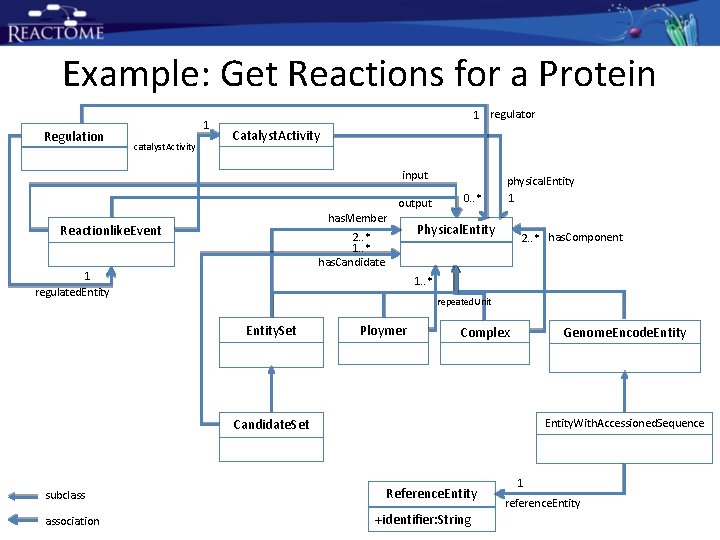
Example: Get Reactions for a Protein Regulation 1 catalyst. Activity 1 regulator Catalyst. Activity input output has. Member Reactionlike. Event 2. . * 1. . * has. Candidate 1 regulated. Entity 0. . * physical. Entity 1 Physical. Entity 2. . * has. Component 1. . * repeated. Unit Entity. Set Ploymer Complex Genome. Encode. Entity. With. Accessioned. Sequence Candidate. Set subclass association Reference. Entity +identifier: String 1 reference. Entity

Example: Get Reactions for a Protein

Example: Get Reactions for a Protein
![Example: Get Reactions for a Protein Reference. Gene. Product: [Reference. Gene. Product: 54208] Uni. Example: Get Reactions for a Protein Reference. Gene. Product: [Reference. Gene. Product: 54208] Uni.](http://slidetodoc.com/presentation_image_h2/c77fd700d7adf291ea2995ba70d00157/image-30.jpg)
Example: Get Reactions for a Protein Reference. Gene. Product: [Reference. Gene. Product: 54208] Uni. Prot: P 00533 EGFR Physical. Entity: 57 [Entity. With. Accessioned. Sequence: 1996343] p-6 Y-EGFR M 766_A 767 ins. ASV mutant [plasma membrane] [Entity. With. Accessioned. Sequence: 1248649] Phospho-EGFRv. IIImut [plasma membrane] [Entity. With. Accessioned. Sequence: 1996341] p-6 Y-EGFR A 289 T mutant [plasma membrane] [Entity. With. Accessioned. Sequence: 1181399] EGFR E 746_A 750 del; T 790 M mutant [plasma membrane] [Entity. With. Accessioned. Sequence: 1177532] EGFR G 719 A mutant [plasma membrane] [Entity. With. Accessioned. Sequence: 1996322] EGFR D 770_N 771 ins. NPG mutant [plasma membrane] [Entity. With. Accessioned. Sequence: 1996326] EGFR M 766_A 767 ins. ASV mutant [plasma membrane] [Entity. With. Accessioned. Sequence: 1169418] EGFR G 719 S mutant [plasma membrane] [Entity. With. Accessioned. Sequence: 1228040] Phospho-EGFR L 747_S 752 del mutant [plasma membrane] …… Reactions: 90 [Reaction: 177944] Activation of SHP 2 through the binding to phospho-Gab 1 [Reaction: 212713] Active PLC-gamma 1 dissociates from EGFR [Reaction: 183077] Assembly in clathrin-coated vesicles (CCVs) [Reaction: 182994] Assembly of EGFR complex in clathrin-coated vesicles [Reaction: 183002] Beta-Pix pushes CIN 85 away from CBL [Reaction: 183055] Binding of CBL to EGFR [Reaction: 1225949] Binding of CBL to phosphorylated ligand-responsive EGFR mutants [Reaction: 1220612] Binding of EGF to ligand-responsive EGFR mutants [Reaction: 1306963] Binding of GRB 2: GAB 1 to p-ERBB 2: p-EGFR ……
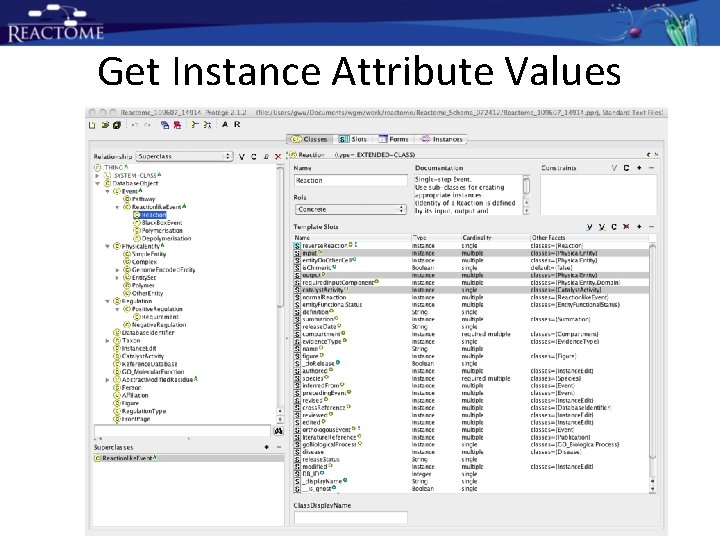
Get Instance Attribute Values

Get Instance Attribute Values
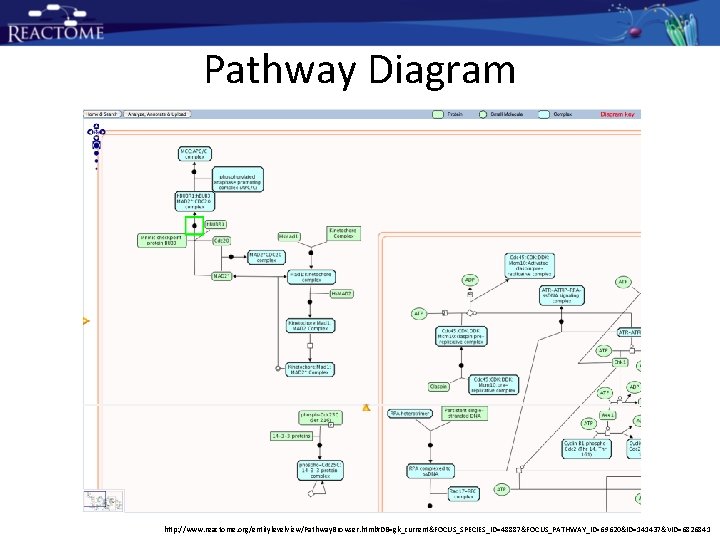
Pathway Diagram http: //www. reactome. org/entitylevelview/Pathway. Browser. html#DB=gk_current&FOCUS_SPECIES_ID=48887&FOCUS_PATHWAY_ID=69620&ID=141437&VID=6826841
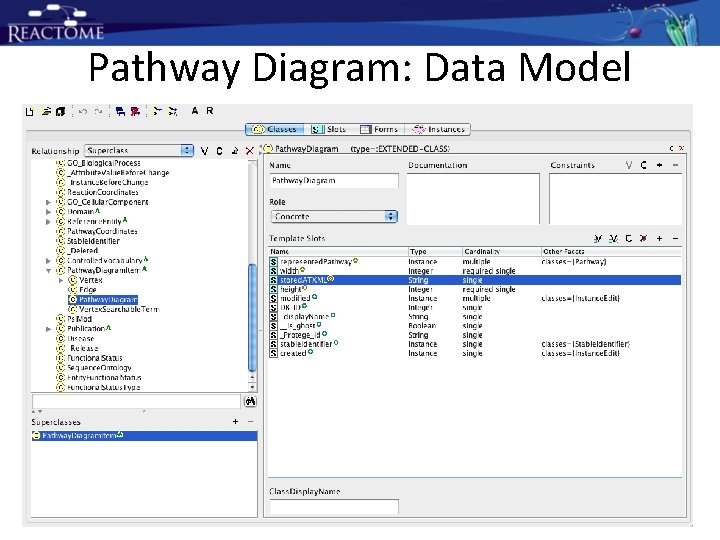
Pathway Diagram: Data Model
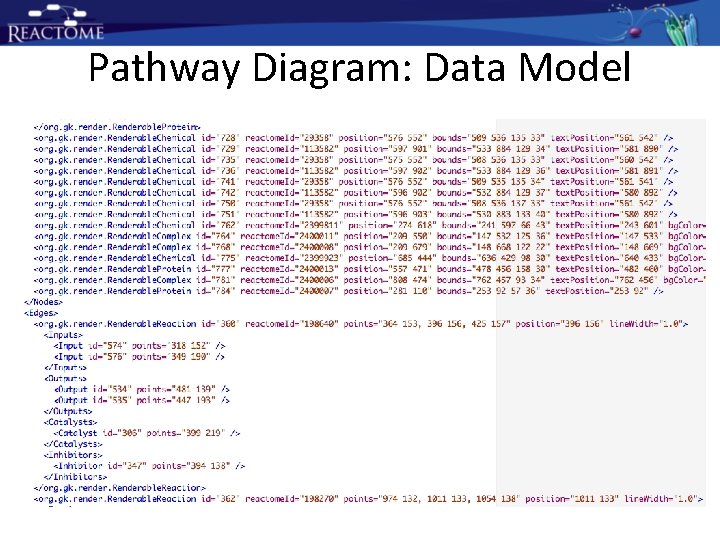
Pathway Diagram: Data Model

Pathway Diagram: Color Protein

Pathway Diagram: Color Protein
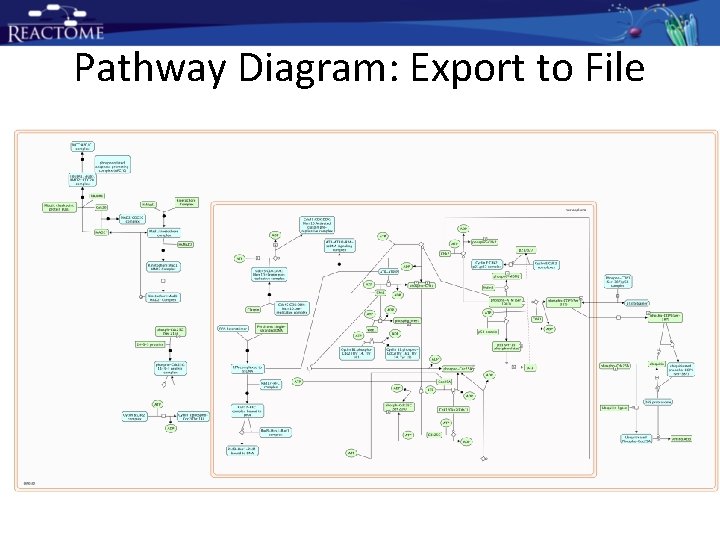
Pathway Diagram: Export to File
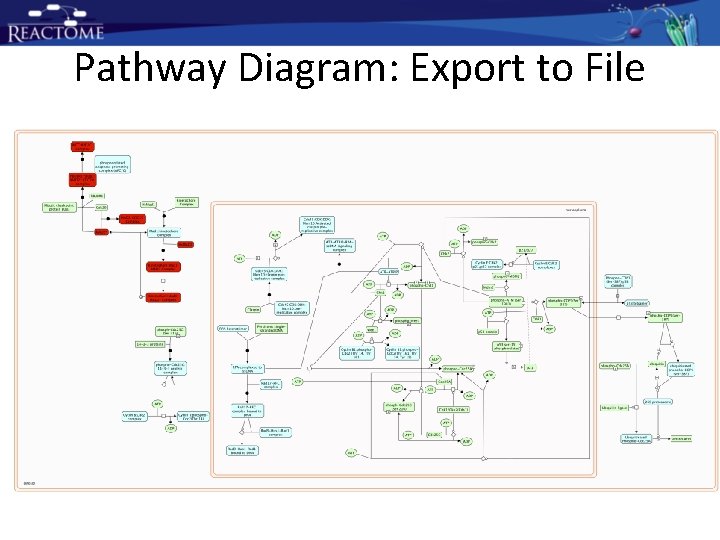
Pathway Diagram: Export to File
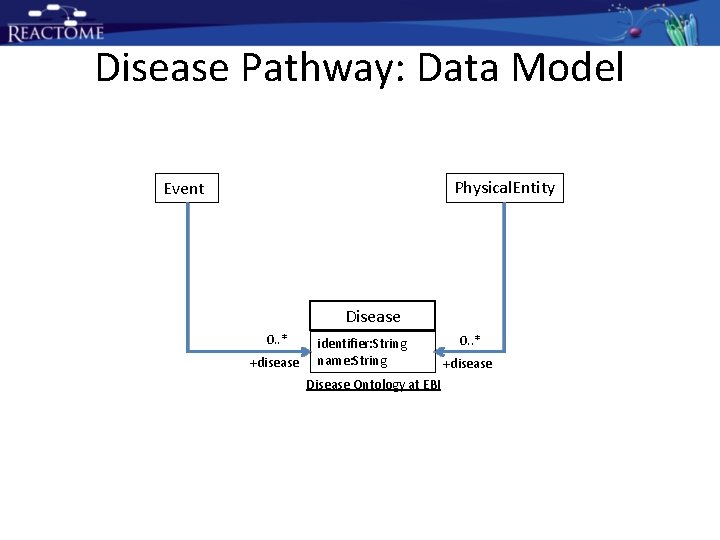
Disease Pathway: Data Model Physical. Entity Event Disease 0. . * +disease identifier: String name: String Disease Ontology at EBI 0. . * +disease
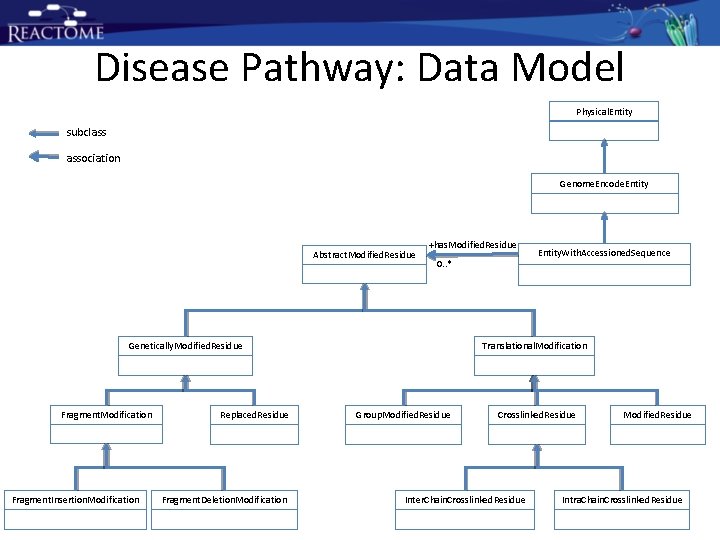
Disease Pathway: Data Model Physical. Entity subclass association Genome. Encode. Entity Abstract. Modified. Residue +has. Modified. Residue 0. . * Translational. Modification Genetically. Modified. Residue Fragment. Modification Fragment. Insertion. Modification Replaced. Residue Fragment. Deletion. Modification Entity. With. Accessioned. Sequence Group. Modified. Residue Crosslinked. Residue Inter. Chain. Crosslinked. Residue Modified. Residue Intra. Chain. Crosslinked. Residue
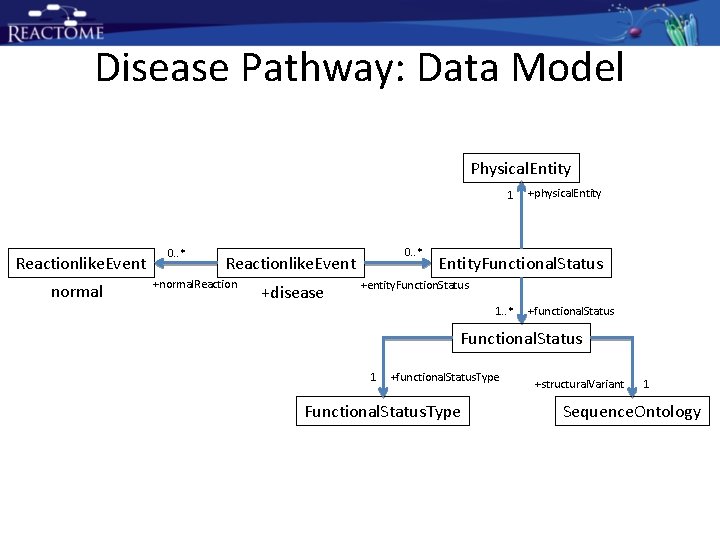
Disease Pathway: Data Model Physical. Entity 1 Reactionlike. Event normal 0. . * Reactionlike. Event +normal. Reaction +disease +physical. Entity. Functional. Status +entity. Function. Status 1. . * +functional. Status Functional. Status 1 +functional. Status. Type Functional. Status. Type +structural. Variant 1 Sequence. Ontology
Disease Pathway: Visualization
Disease Pathway: Visualization
Disease Pathway: Visualization
Disease Pathway: Visualization
Disease Pathway: Visualization
Perl API
Perl API http: //www. reactome. org/download/index. html
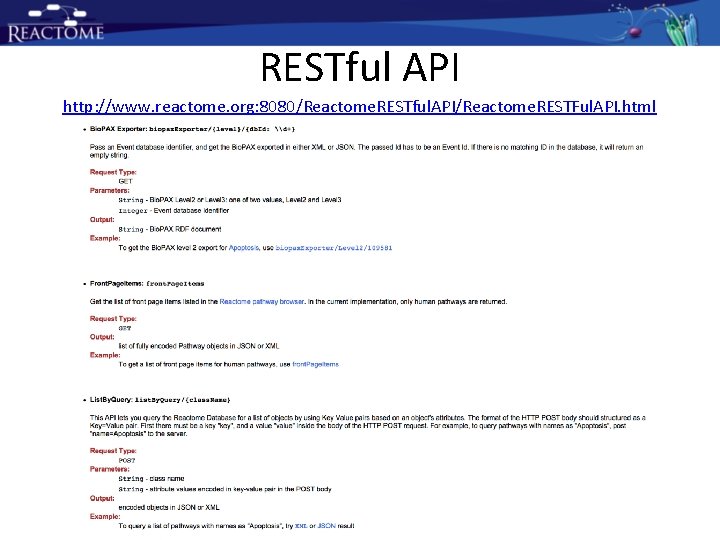
RESTful API http: //www. reactome. org: 8080/Reactome. RESTful. API/Reactome. RESTFul. API. html
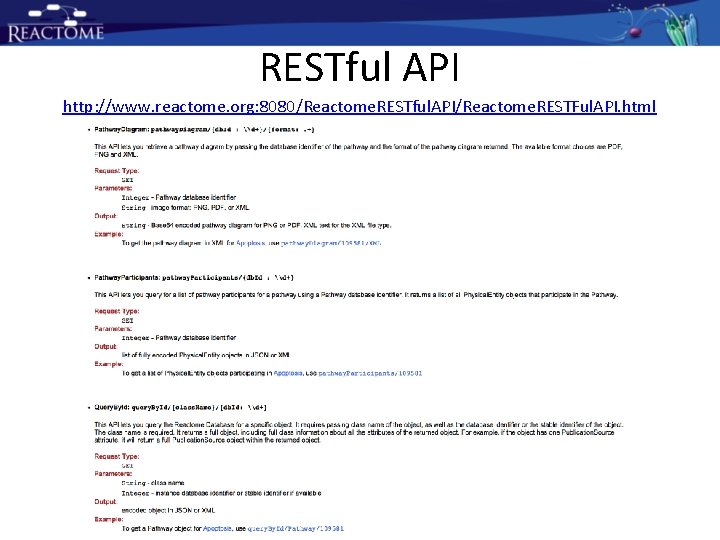
RESTful API http: //www. reactome. org: 8080/Reactome. RESTful. API/Reactome. RESTFul. API. html
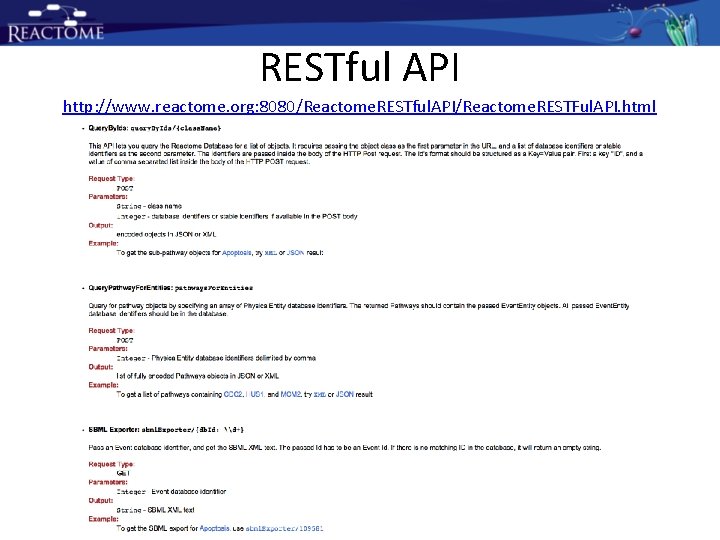
RESTful API http: //www. reactome. org: 8080/Reactome. RESTful. API/Reactome. RESTFul. API. html

SOAP API http: //www. reactome. org: 8080/ca. BIOWeb. App/docs/services. html

SOAP API http: //www. reactome. org: 8080/ca. BIOWeb. App/docs/ca. BIG_Reactome_User_Guide. pdf
Acknowledgements • Java and Perl API Imre Vastrik David Croft • RESTful API Stanislav Palatnik (GSo. C) • Disease model Marija Orlic-Milacic Karen Rothfels Robin Haw Lisa Matthew Bijay Jassai Heeyeon Song Marc Gillespie Steve Jupe Peter D’Eustachio
- Slides: 55