About OMICS Group is an amalgamation of Open

About OMICS Group is an amalgamation of Open Access publications and worldwide international science conferences and events. Established in the year 2007 with the sole aim of making the information on Sciences and technology ‘Open Access’, OMICS Group publishes 500 online open access scholarly journals in all aspects of Science, Engineering, Management and Technology journals. OMICS Group has been instrumental in taking the knowledge on Science & technology to the doorsteps of ordinary men and women. Research Scholars, Students, Libraries, Educational Institutions, Research centers and the industry are main stakeholders that benefitted greatly from this knowledge dissemination. OMICS Group also organizes 500 International conferences annually across the globe, where knowledge transfer takes place through debates, round table discussions, poster presentations, workshops, symposia and exhibitions.

About OMICS International Conferences OMICS International is a pioneer and leading science event organizer, which publishes around 500 open access journals and conducts over 500 Medical, Clinical, Engineering, Life Sciences, Pharma scientific conferences all over the globe annually with the support of more than 1000 scientific associations and 30, 000 editorial board members and 3. 5 million followers to its credit. OMICS Group has organized 500 conferences, workshops and national symposiums across the major cities including San Francisco, Las Vegas, San Antonio, Omaha, Orlando, Raleigh, Santa Clara, Chicago, Philadelphia, Baltimore, United Kingdom, Valencia, Dubai, Beijing, Hyderabad, Bengaluru and Mumbai. .

Dr. A. P. J. Abdul Kalam (15 October 1931 – 27 July 2015)

De Novo Transcriptome Assembly and Identification of Cold and Freeze Responsive Genes in Seabuckthorn. Saurabh Chaudhary and Prakash Chand Sharma University School of Biotechnology, Guru Gobind Singh Indraprastha University, Sector -16 C, Dwarka, New Delhi. 110078

Seabuckthorn • Hardy, deciduous shrub (family Elaeagnaceae) • Yellow or orange berries have tremendous medicinal & nutritional values • Indian Himalayas 2 nd richest source in world • Prevents soil erosion and helps in land reclamation • Grows in extreme environments: important source of new genes (alleles) for abiotic stress tolerance
Objectives 1. Seabuckthorn transcriptome assembly and analysis using NGS 2. Deep. SAGE based identification of differentially expressed genes during cold and freeze stress 3. Validation of selected cold and freeze responsive genes using q. RT-PCR
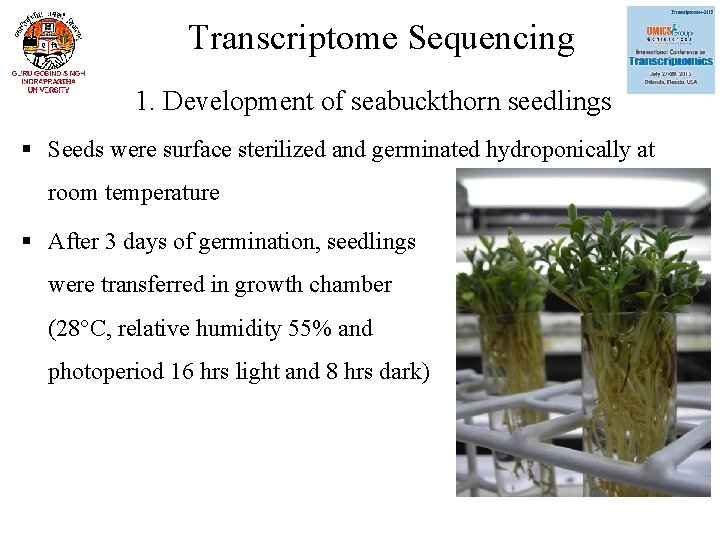
Transcriptome Sequencing 1. Development of seabuckthorn seedlings § Seeds were surface sterilized and germinated hydroponically at room temperature § After 3 days of germination, seedlings were transferred in growth chamber (28°C, relative humidity 55% and photoperiod 16 hrs light and 8 hrs dark)
2. RNA isolation and quality control § Total RNA isolated using modified CTAB method with Invisorb Invitek RNA isolation columns § Good quality and high yield of total RNA was obtained from leaf and root tissues § The RNA samples with A 260/A 280 ratio from 1. 9 to 2. 1, A 260/A 230 ratio from 2. 0 to 2. 5 and RIN value more than 8. 0 were further processed for next generation sequencing
3. Transcriptome Assembly § High throughput next generation sequencing (Illumina Solexa) platform was used for sequencing transcripts § Raw and filtered short read data (NGS QC Tool kit) Item Leaf Root Total number of reads 44791536 49222400 Total number of HQ reads 41067522 45186352 91. 69% 91. 80% Percentage of HQ reads

§ Approx. 94 million short reads generated § Submitted to NCBI-SRA vide Accession No. SRA 050428 § Approx. 86 million reads of high quality were further taken for assembly § De novo assemblers used to optimize transcriptome assembly Ø Velvet Ø Oases Ø ABy. SS Ø SOAPdenovo Ø CLC Genomics Workbench Ø Trinity
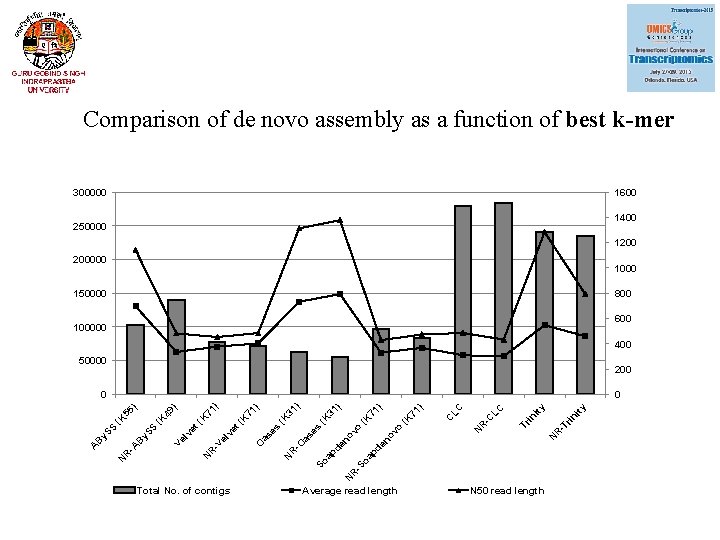
Comparison of de novo assembly as a function of best k-mer 300000 1600 1400 250000 1200 200000 150000 800 600 100000 400 50000 200 Total No. of contigs Average read length ity rin in -T Tr N R -C LC LC N R -S oa pd C ) 71 (K en ov o en o ap d So N R -O as as e vo s es (K (K 71 ) 31 ) (K 71 ) N R -V el ve t( K K et ( lv Ve (K y. S S B -A R N O ) 49 55 (K S B y. S A 71 ) 0 N 50 read length
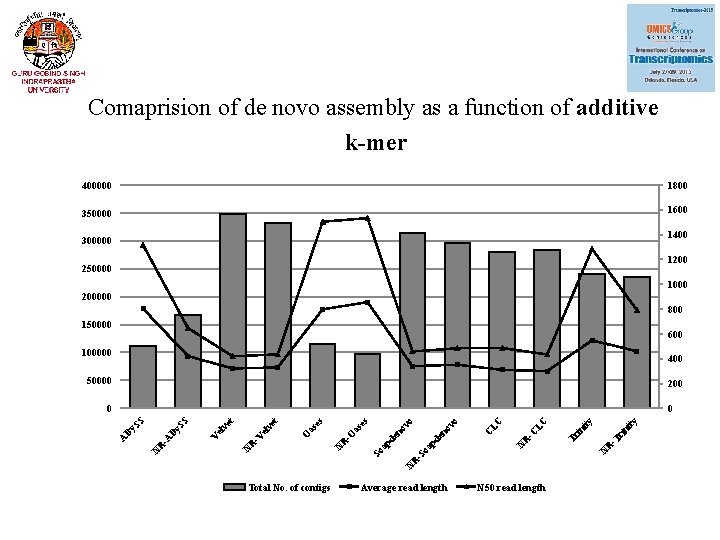
Comaprision of de novo assembly as a function of additive k-mer 400000 1800 350000 1600 1400 300000 1200 250000 1000 200000 800 150000 600 100000 400 50000 200 Total No. of contigs Average read length ni ty ity R -T ri in Tr N N R -C LC LC en ov pd N R -S oa C o o pd So a -O R N en ov as es O et el v R -V N V el SS N R -A By By A ve t 0 SS 0 N 50 read length
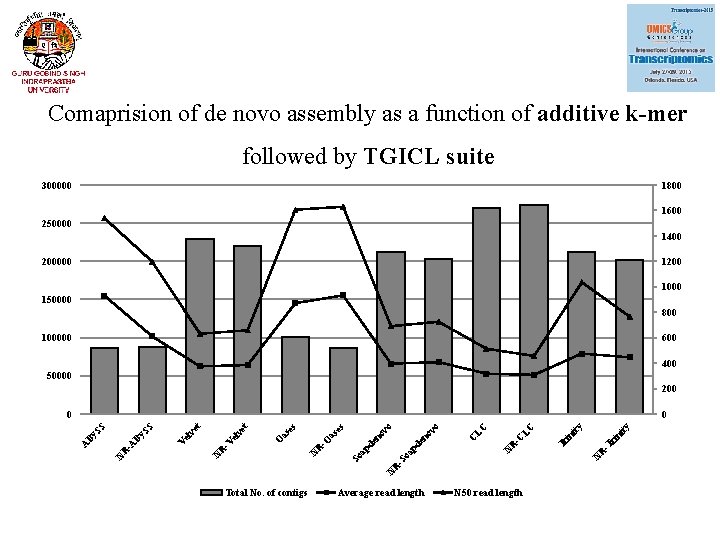
Comaprision of de novo assembly as a function of additive k-mer followed by TGICL suite 300000 1800 1600 250000 1400 200000 1200 1000 150000 800 100000 600 400 50000 200 Total No. of contigs Average read length N 50 read length ni ty ri -T Tr in ity N R -C LC LC en ov pd N R -S oa C o o pd So a -O R N en ov as es O et N R -V el v V SS N R -A By By A et 0 SS 0
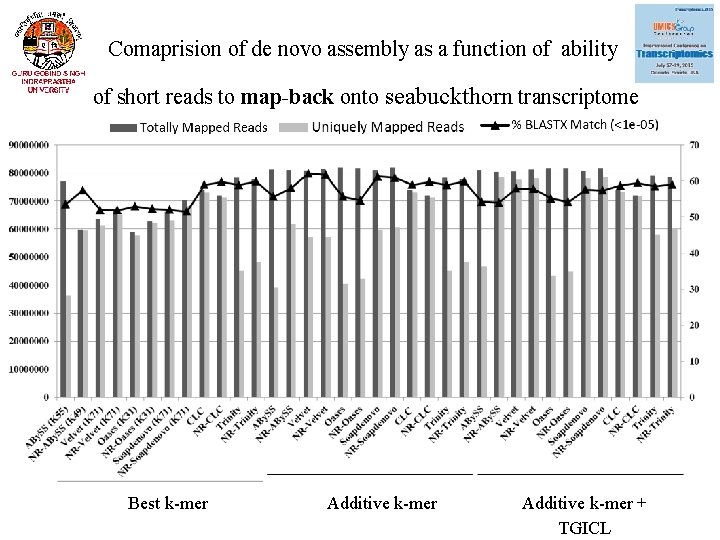
Comaprision of de novo assembly as a function of ability of short reads to map-back onto seabuckthorn transcriptome Best k-mer Additive k-mer + TGICL
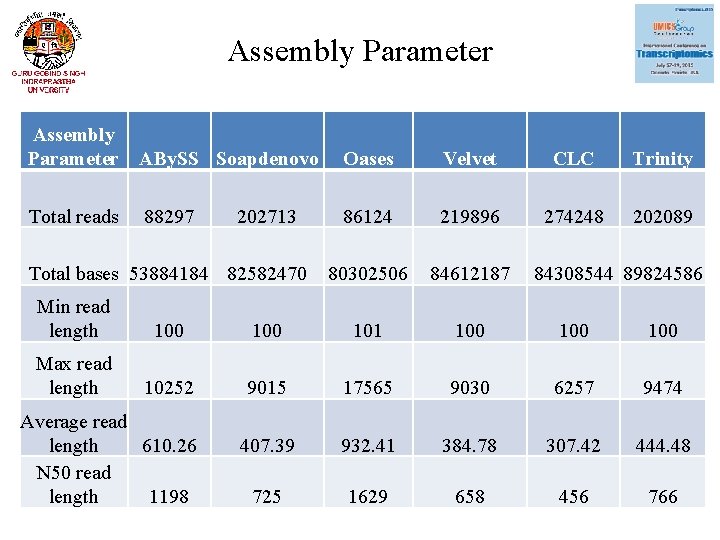
Assembly Parameter ABy. SS Soapdenovo Oases Velvet CLC Trinity Total reads 86124 219896 274248 202089 80302506 84612187 88297 202713 Total bases 53884184 82582470 84308544 89824586 Min read length 100 101 100 100 Max read length 10252 9015 17565 9030 6257 9474 407. 39 932. 41 384. 78 307. 42 444. 48 725 1629 658 456 766 Average read length 610. 26 N 50 read length 1198
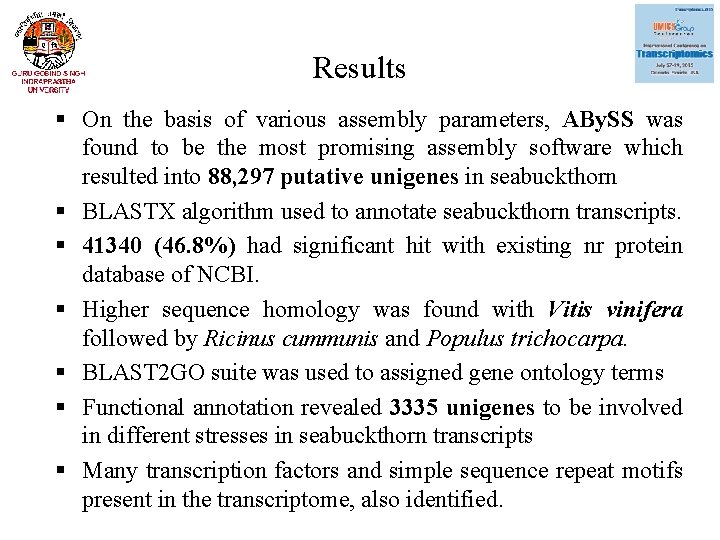
Results § On the basis of various assembly parameters, ABy. SS was found to be the most promising assembly software which resulted into 88, 297 putative unigenes in seabuckthorn § BLASTX algorithm used to annotate seabuckthorn transcripts. § 41340 (46. 8%) had significant hit with existing nr protein database of NCBI. § Higher sequence homology was found with Vitis vinifera followed by Ricinus cummunis and Populus trichocarpa. § BLAST 2 GO suite was used to assigned gene ontology terms § Functional annotation revealed 3335 unigenes to be involved in different stresses in seabuckthorn transcripts § Many transcription factors and simple sequence repeat motifs present in the transcriptome, also identified.
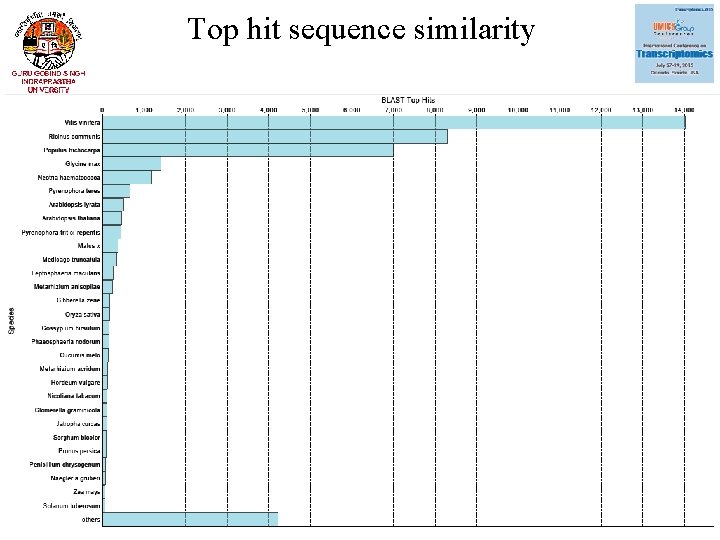
Top hit sequence similarity
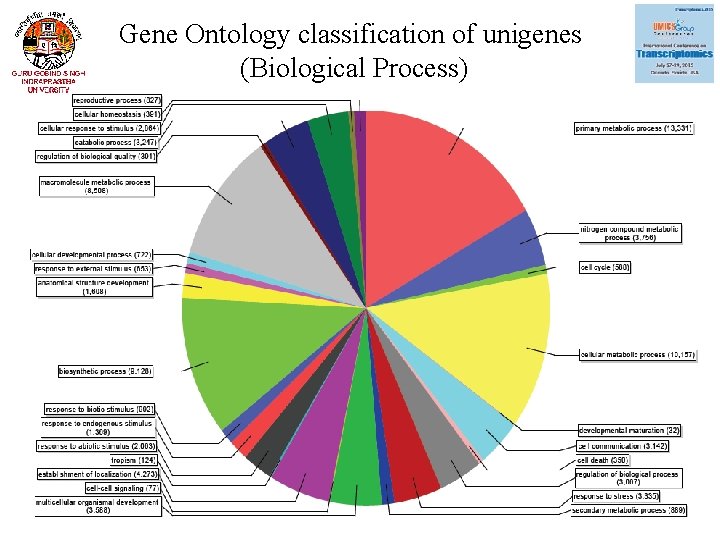
Gene Ontology classification of unigenes (Biological Process)
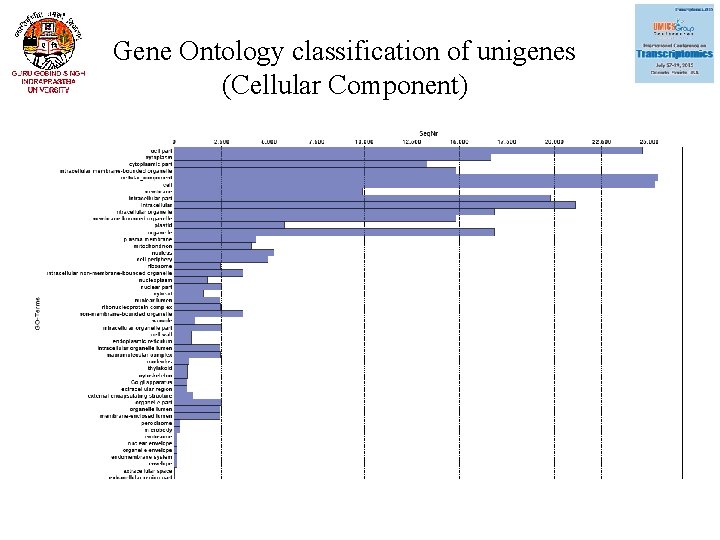
Gene Ontology classification of unigenes (Cellular Component)
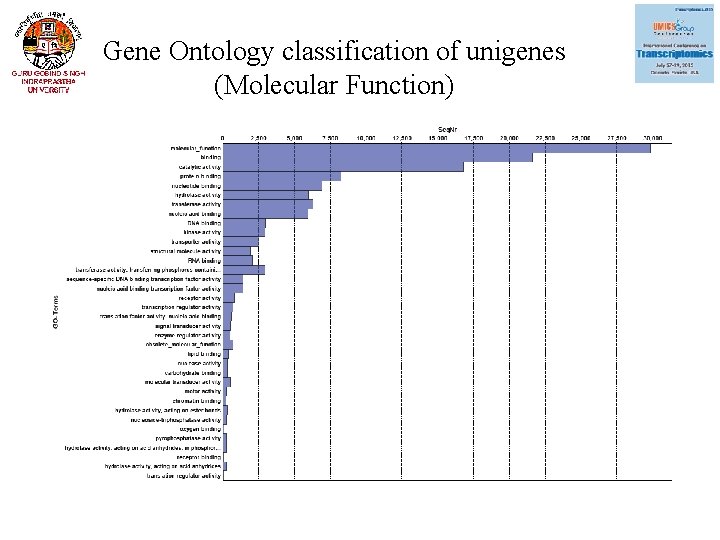
Gene Ontology classification of unigenes (Molecular Function)

Deep. SAGE • A combination of classical SAGE (Serial Analysis of Gene Expression) and Next generation Sequencing, used for global gene expression analysis under certain environment • Also known as Digital Gene Expression (DGE) Profiling
Digital Gene Expression Profiling (Deep. SAGE) § 30 days old seabuckthorn seedlings were subjected to cold treatment at 4°C and freeze treatment at -10°C for 6 hours § Seedlings grown at 26°C were taken as control § Leaf tissues were harvested from all three setups § Total RNA isolation and quality check from all three samples § The 17 base pair tag were generated as per manufacturer protocol for Deep. SAGE
Digital gene expression (Sequencing and Analysis) § Tags were directly sequenced using next generation sequencing (Illumina Solexa) platform § Analysis of tags generated revealed large number differentially expressed genes in response to cold and freeze stress. § Differentially expressed genes were annotated by sequence similarity search (BLAST) with our existing seabuckthorn transcriptome § Around 450 genes identified responsive to cold and freeze stress in seabuckthorn.
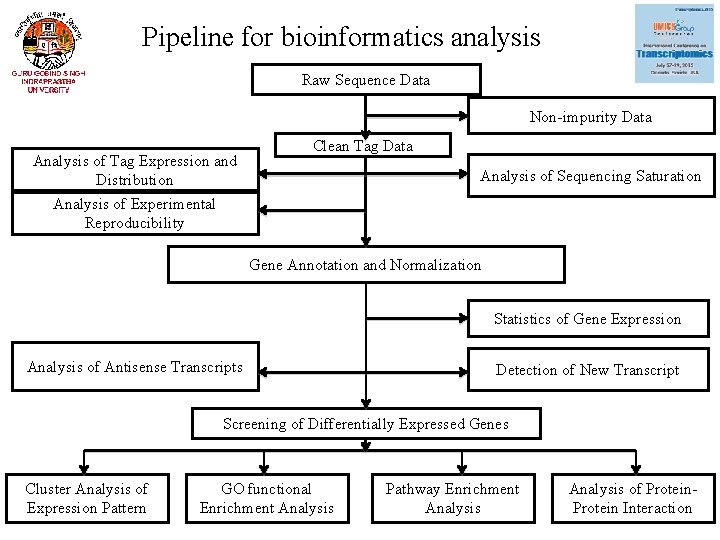
Pipeline for bioinformatics analysis Raw Sequence Data Non-impurity Data Analysis of Tag Expression and Distribution Clean Tag Data Analysis of Sequencing Saturation Analysis of Experimental Reproducibility Gene Annotation and Normalization Statistics of Gene Expression Analysis of Antisense Transcripts Detection of New Transcript Screening of Differentially Expressed Genes Cluster Analysis of Expression Pattern GO functional Enrichment Analysis Pathway Enrichment Analysis of Protein Interaction
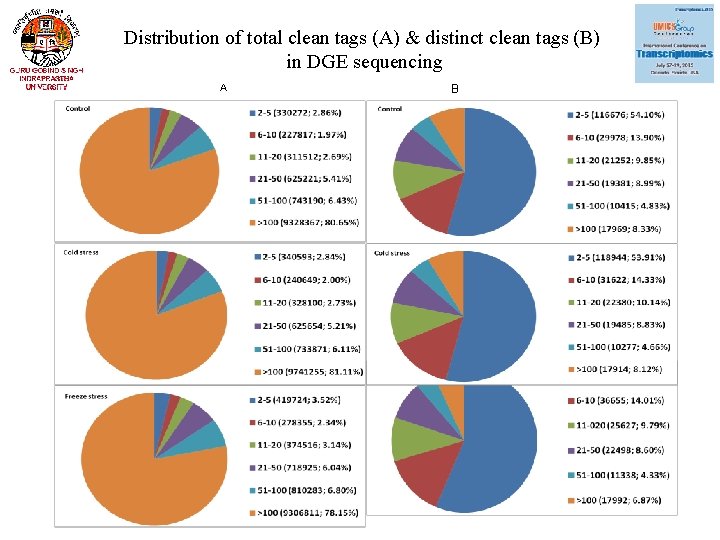
Distribution of total clean tags (A) & distinct clean tags (B) in DGE sequencing A B
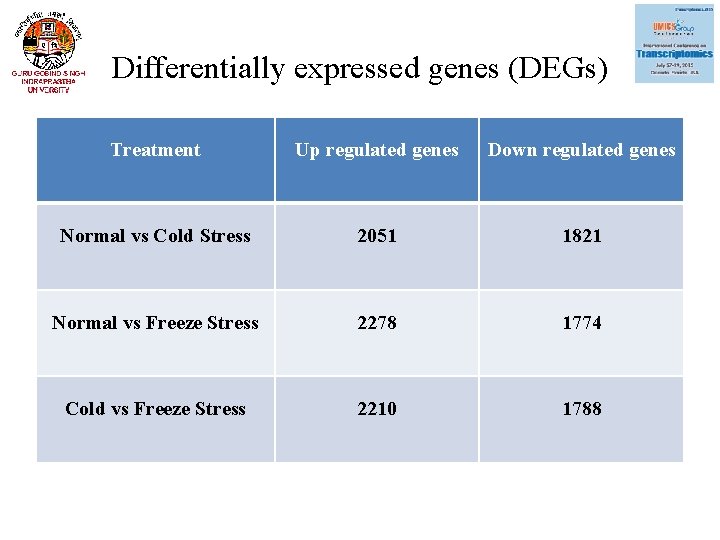
Differentially expressed genes (DEGs) Treatment Up regulated genes Down regulated genes Normal vs Cold Stress 2051 1821 Normal vs Freeze Stress 2278 1774 Cold vs Freeze Stress 2210 1788
Gene ontology and KEGG pathway analysis § GO- terms suggested metabolic, cellular, primary metabolic, cellular metabolic and macromolecule metabolic processes genes were found to be abundant under cold and freeze stress § 28% and 15% of the differentially expressed genes were categorized as genes responsive to stimulus and stress, respectively § Photosynthetic, carotenoid biosynthesis and plant hormone signal transduction and vitamin B 6 metabolism pathways found to be the most significantly enriched pathways during cold and freeze stress in seabuckthorn.
Validation of selected genes (Quantitative real-time RT-PCR) § Differentially expressed genes BLAST with existing annotated unigenes of seabuckthorn § 22 genes from Deep. SAGE data selected (involved in cold and freeze stress for validation) § The seabuckthorn seedlings were given various cold and freeze treatments at different time interval § Real time expression: biological duplicate and technical triplicates for each samples
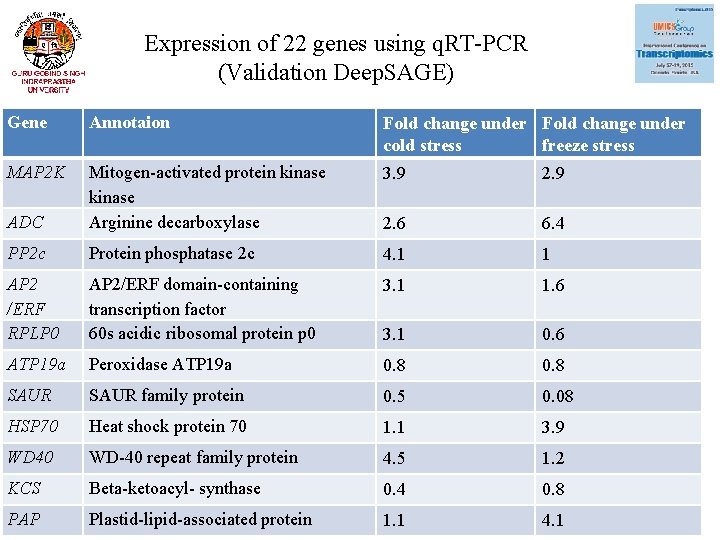
Expression of 22 genes using q. RT-PCR (Validation Deep. SAGE) Gene Annotaion Fold change under cold stress freeze stress MAP 2 K 3. 9 2. 9 ADC Mitogen-activated protein kinase Arginine decarboxylase 2. 6 6. 4 PP 2 c Protein phosphatase 2 c 4. 1 1 AP 2 /ERF RPLP 0 AP 2/ERF domain-containing transcription factor 60 s acidic ribosomal protein p 0 3. 1 1. 6 3. 1 0. 6 ATP 19 a Peroxidase ATP 19 a 0. 8 SAUR family protein 0. 5 0. 08 HSP 70 Heat shock protein 70 1. 1 3. 9 WD 40 WD-40 repeat family protein 4. 5 1. 2 KCS Beta-ketoacyl- synthase 0. 4 0. 8 PAP Plastid-lipid-associated protein 1. 1 4. 1
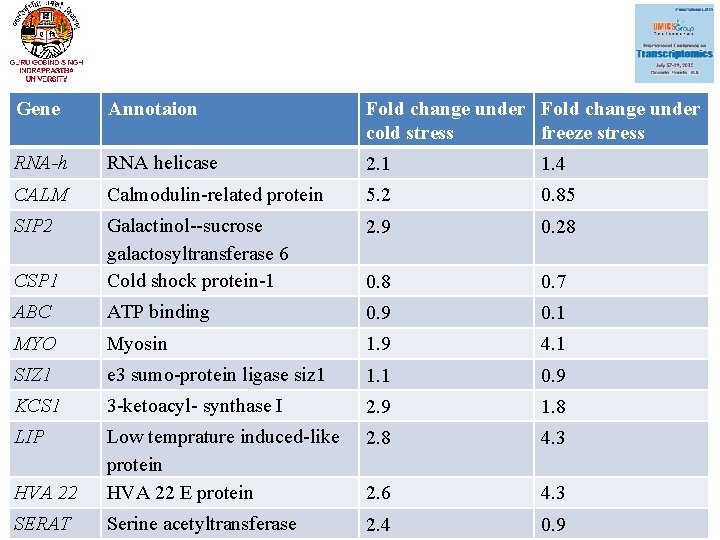
Gene Annotaion Fold change under cold stress freeze stress RNA-h RNA helicase 2. 1 1. 4 CALM Calmodulin-related protein 5. 2 0. 85 SIP 2 2. 9 0. 28 CSP 1 Galactinol--sucrose galactosyltransferase 6 Cold shock protein-1 0. 8 0. 7 ABC ATP binding 0. 9 0. 1 MYO Myosin 1. 9 4. 1 SIZ 1 e 3 sumo-protein ligase siz 1 1. 1 0. 9 KCS 1 3 -ketoacyl- synthase I 2. 9 1. 8 LIP 2. 8 4. 3 HVA 22 Low temprature induced-like protein HVA 22 E protein 2. 6 4. 3 SERAT Serine acetyltransferase 2. 4 0. 9
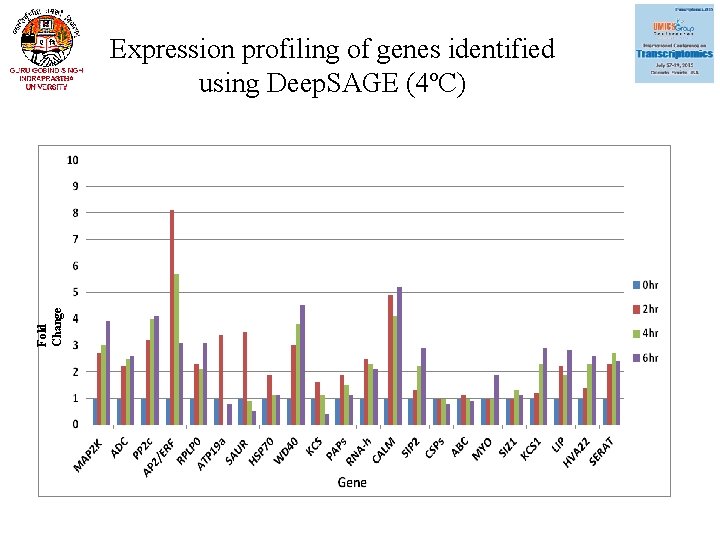
Fold Change Expression profiling of genes identified using Deep. SAGE (4ºC)
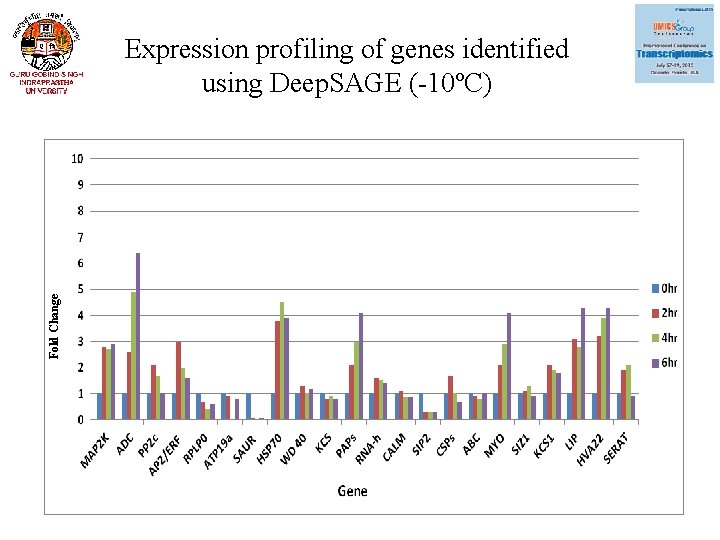
Fold Change Expression profiling of genes identified using Deep. SAGE (-10ºC)

Conclusions • Standardization of high quality RNA isolation from seabuckthorn and establishment of seabuckthorn seedlings in laboratory • Whole transcriptome data obtained using next generation sequencing platform provides an upper edge as reference dataset for further studies • Unigenes with no relevant hit in NCBI nr protein database were considered to be seabuckthorn specific and can further be characterized in future
• Standardization of assembly tools for next generation sequencing data helps further in analysis of NGS data for other species • Identification of 7421 putative seabuckthorn transcription factor genes • Development of USMM for seabuckthorn • Many abiotic stress responsive genes, specifically for cold and freeze tolerant in seabuckthorn have been identified.

• Validation of 20 most important expressed genes during cold and freeze stress, revealed active expression of cold/freeze stress responsible genes in seabuckthorn.

Research Leads • Genes identified to be involved in cold tolerance need to be further characterized • Some of these genes be explored for development of cold tolerant transgenic • Assembler (ABy. SS) optimized for NGS assembly can be used for other similar studies

Acknowledgement • Department of Biotechnology, Government of India • ITS- Department of Science and Technology Government of India • Guru Gobind Singh Indraprastha University, India • Omics International Conferences

Thank You

Let Us Meet Again We welcome you all to our future conferences of OMICS International Please Visit: http: //transcriptomics. conferenceseries. com/ http: //www. conferenceseries. com/genetics-and-molecularbiology-conferences. php
- Slides: 41