A Tutorial on Neural Network Autoencoder Yimin Hu











- Slides: 45

A Tutorial on Neural Network & Autoencoder Yimin Hu Quon Lab UC Davis Genome Center July 29, 2016

Outline A Background B Neural Network C Example: Connectivity. Map Contest D Autoencoder E Summary

Part 1 Background of Machine Learing

What can machine learning do?

Why do we need machine learning in gene data/ bioinformatics? Ø Scale of the data has become much larger than before. Ø Gene expression for every single gene is not independent, often there will be some (non-linear) relationship between each other. Ø Both can be solved by machine learning algorithms

Basic notion of Machine (Statistical) Learning Ø Given a dataset which has input vector x, which we often call the vector of “features”, and output y which stands for labels of classes or a real value. Ø What Machine Learning algorithms do is figure out the function ; it can either be a regression problem or a classification problem. Ø We choose a model and train the model with the given data, then we will be able to predict the output for new coming data. Ø Supervised: Training with labeled data Ø Unsupervised: Training with unlabeled data

Part 2 Neural Network

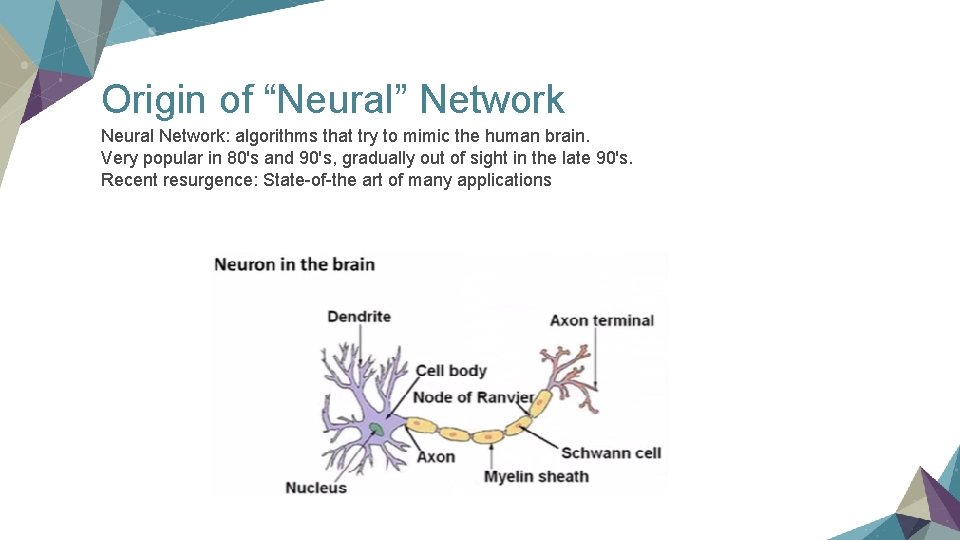
Origin of “Neural” Network Neural Network: algorithms that try to mimic the human brain. Very popular in 80's and 90's, gradually out of sight in the late 90's. Recent resurgence: State-of-the art of many applications

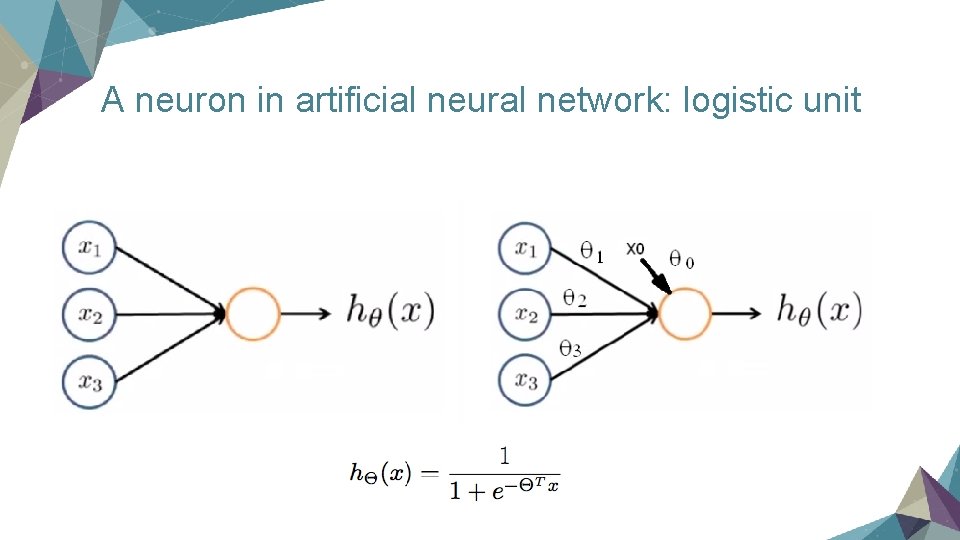
A neuron in artificial neural network: logistic unit

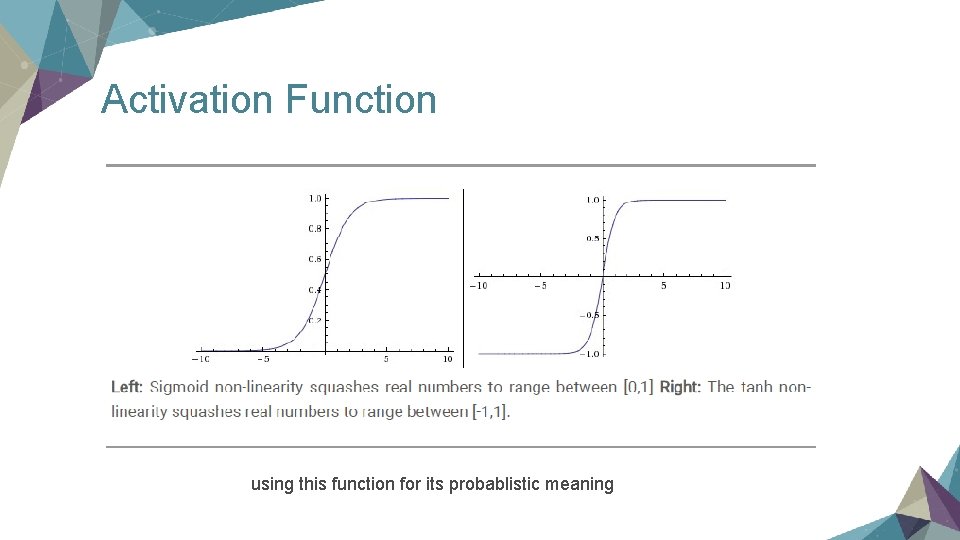
Activation Function using this function for its probablistic meaning

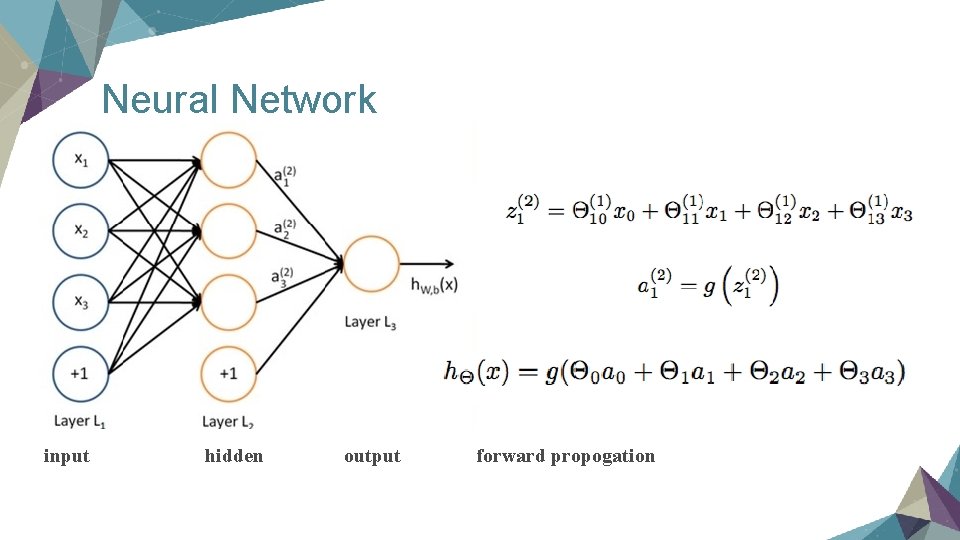
Neural Network input hidden output forward propogation

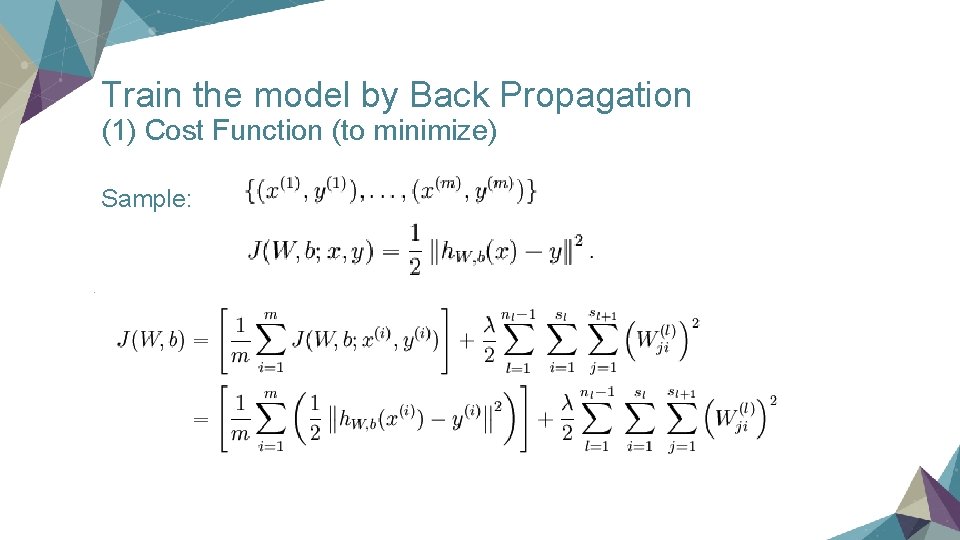
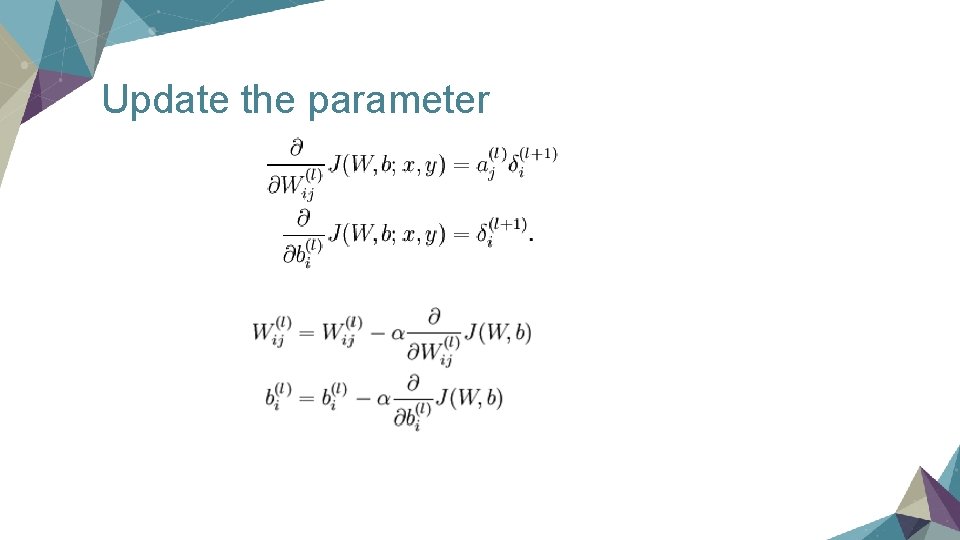
Train the model by Back Propagation (1) Cost Function (to minimize) Sample:

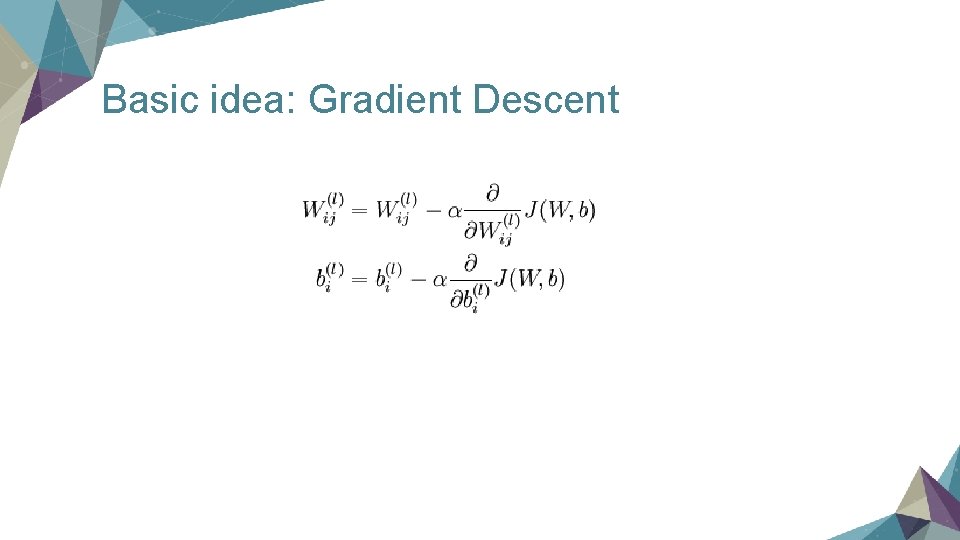
Basic idea: Gradient Descent

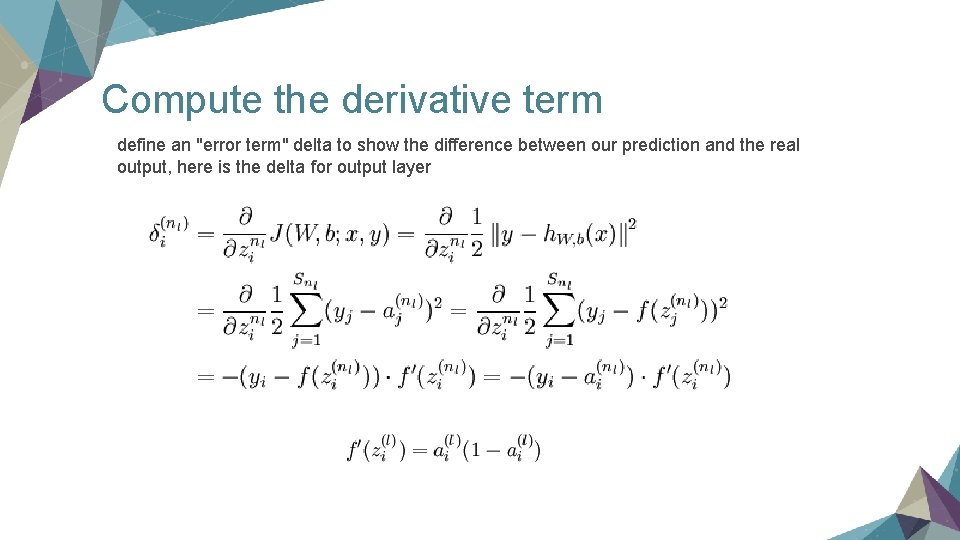
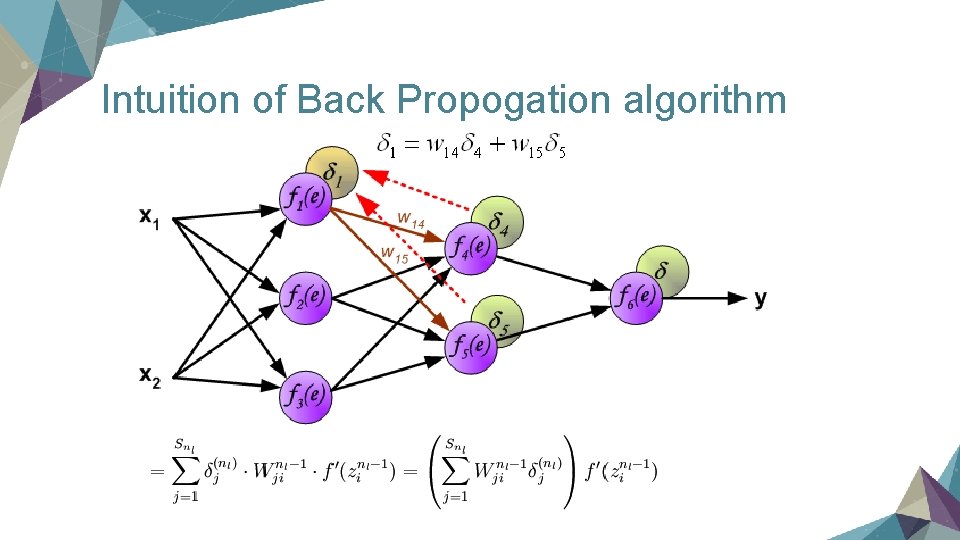
Compute the derivative term define an "error term" delta to show the difference between our prediction and the real output, here is the delta for output layer

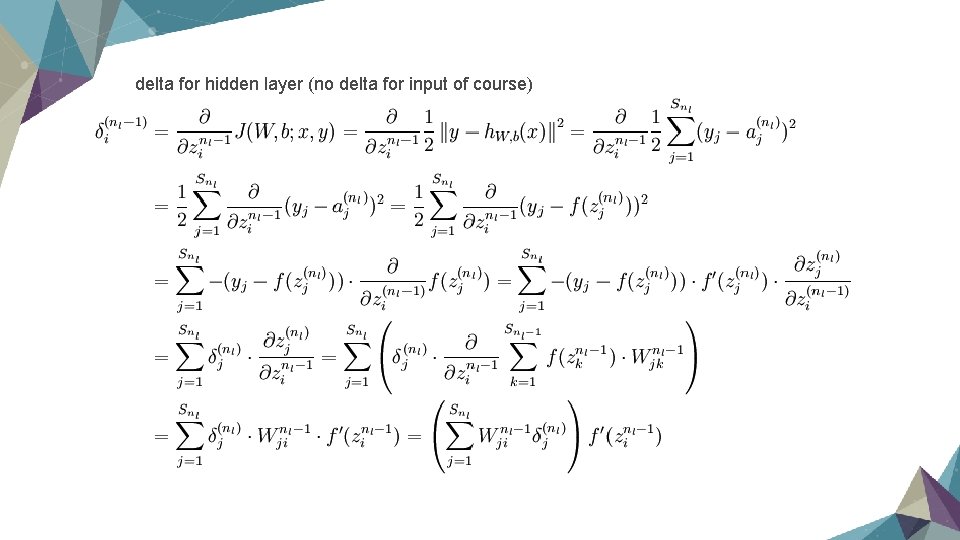
delta for hidden layer (no delta for input of course)

Intuition of Back Propogation algorithm

Update the parameter

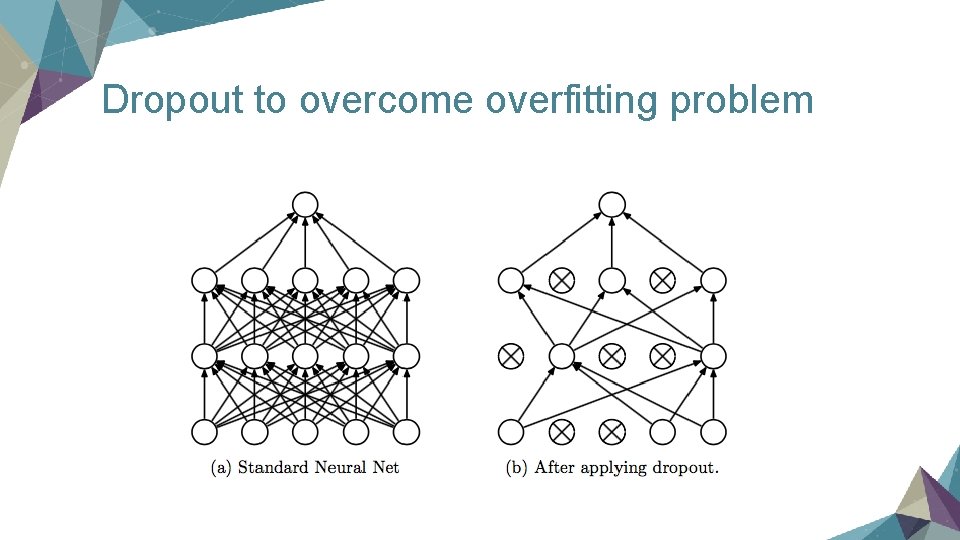
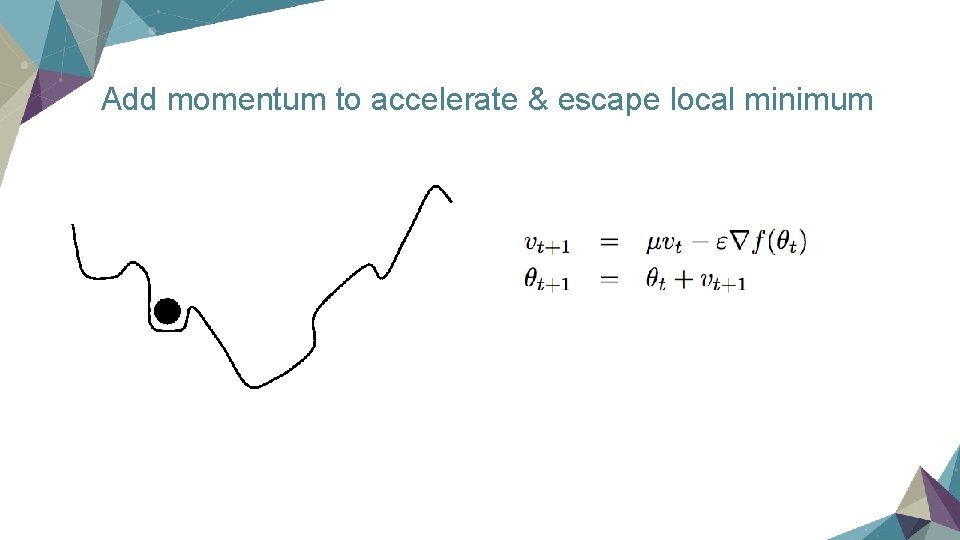
Key points in training a neural network Ø Dropout Ø Momentum

Dropout to overcome overfitting problem

Add momentum to accelerate & escape local minimum

Part 3 Example: Connectivity. Map Contest

Background of CMap Data Ø New technology makes it possible to get the gene profile at scale, but it will cost a lot to test all the genes. Ø The crux of CMap is that it tries to use only about 1000 genes' data to predict the rest of the genes (~20000 genes in human cell) Ø Can be done because some genes’ expression levels are correlated via functional redundancy and/or shared regulatory mechanisms

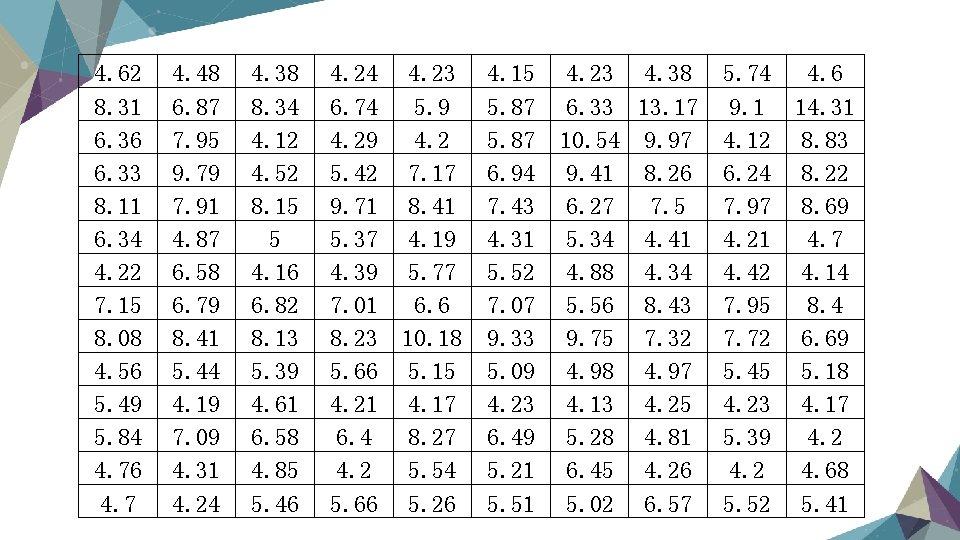
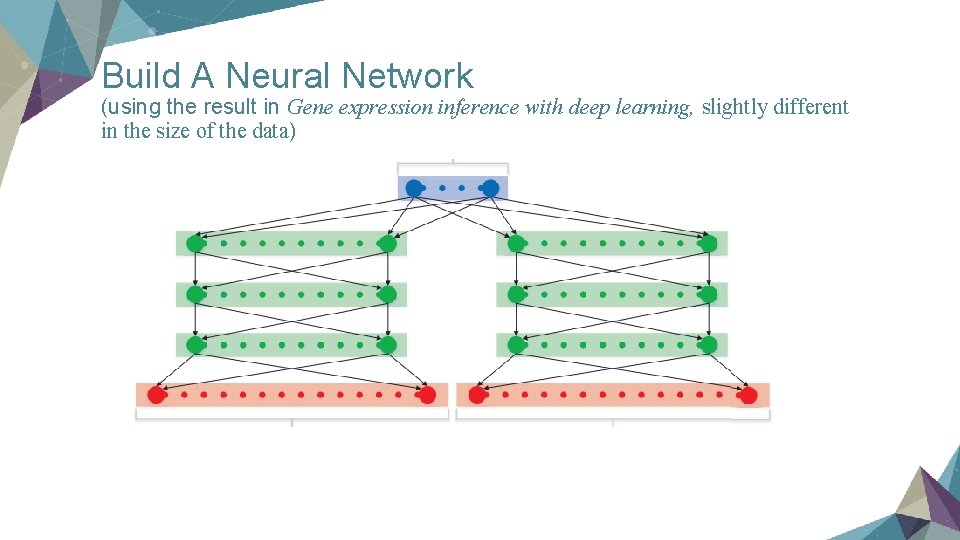
Data Format Ø Basically we have data as a matrix, each row stands for a single gene expression, each column stands for a sample. Ø The meaning of each sample is: we apply 20000 kinds of drug to 5 different kinds of tissue to see how they effect the expression of gene. So we have 100000 samples as our training data. Ø The real value in the matrix is log(gene intensity), which are in the range of 0. 00 -15. 00.

4. 62 8. 31 6. 36 6. 33 8. 11 6. 34 4. 22 7. 15 8. 08 4. 56 5. 49 5. 84 4. 76 4. 7 4. 48 6. 87 7. 95 9. 79 7. 91 4. 87 6. 58 6. 79 8. 41 5. 44 4. 19 7. 09 4. 31 4. 24 4. 38 8. 34 4. 12 4. 52 8. 15 5 4. 16 6. 82 8. 13 5. 39 4. 61 6. 58 4. 85 5. 46 4. 24 6. 74 4. 29 5. 42 9. 71 5. 37 4. 39 7. 01 8. 23 5. 66 4. 21 6. 4 4. 2 5. 66 4. 23 5. 9 4. 2 7. 17 8. 41 4. 19 5. 77 6. 6 10. 18 5. 15 4. 17 8. 27 5. 54 5. 26 4. 15 5. 87 6. 94 7. 43 4. 31 5. 52 7. 07 9. 33 5. 09 4. 23 6. 49 5. 21 5. 51 4. 23 4. 38 6. 33 13. 17 10. 54 9. 97 9. 41 8. 26 6. 27 7. 5 5. 34 4. 41 4. 88 4. 34 5. 56 8. 43 9. 75 7. 32 4. 98 4. 97 4. 13 4. 25 5. 28 4. 81 6. 45 4. 26 5. 02 6. 57 5. 74 9. 1 4. 12 6. 24 7. 97 4. 21 4. 42 7. 95 7. 72 5. 45 4. 23 5. 39 4. 2 5. 52 4. 6 14. 31 8. 83 8. 22 8. 69 4. 7 4. 14 8. 4 6. 69 5. 18 4. 17 4. 2 4. 68 5. 41

Purpose of the CMap Contest Ø using 970 landmark genes to predict other 11350 genes Ø just a regression problem, so we can use any statistical model Ø linear regression is effective but imperfect

Build A Neural Network (using the result in Gene expression inference with deep learning, slightly different in the size of the data)

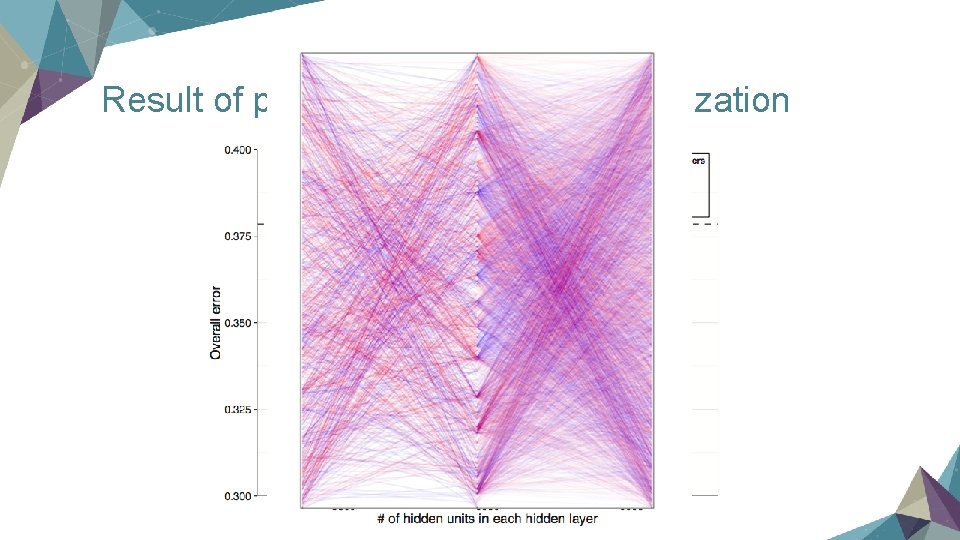
Result of prediction & Network visualization

Conclusion Ø Neural Network is helpful to retrieve the inner relationship among genes and if we choose the landmarks correctly, they will have very strong power to predict the rest of other genes. Ø It's really a powerful tool!

Part 4 Autoencoder

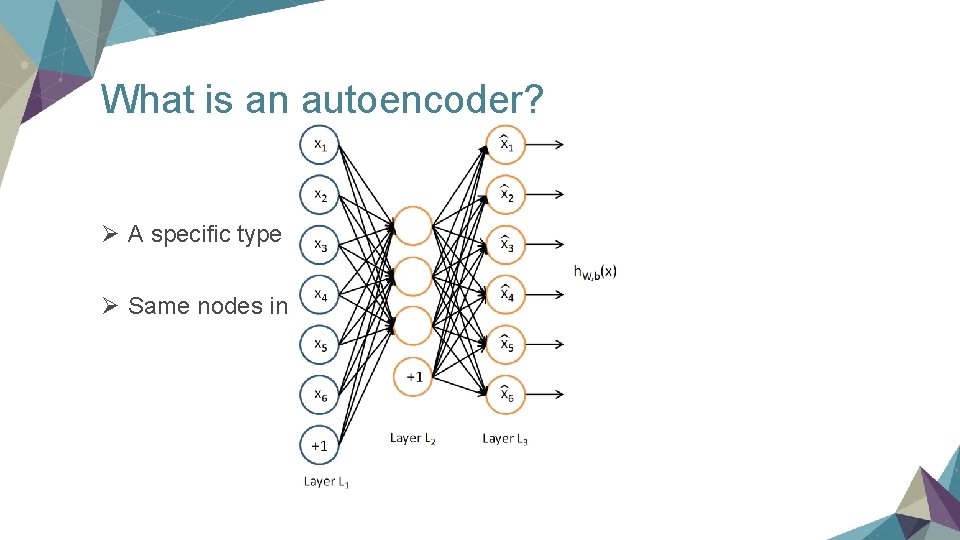
What is an autoencoder? Ø A specific type of NN Ø Same nodes in the input and output layer

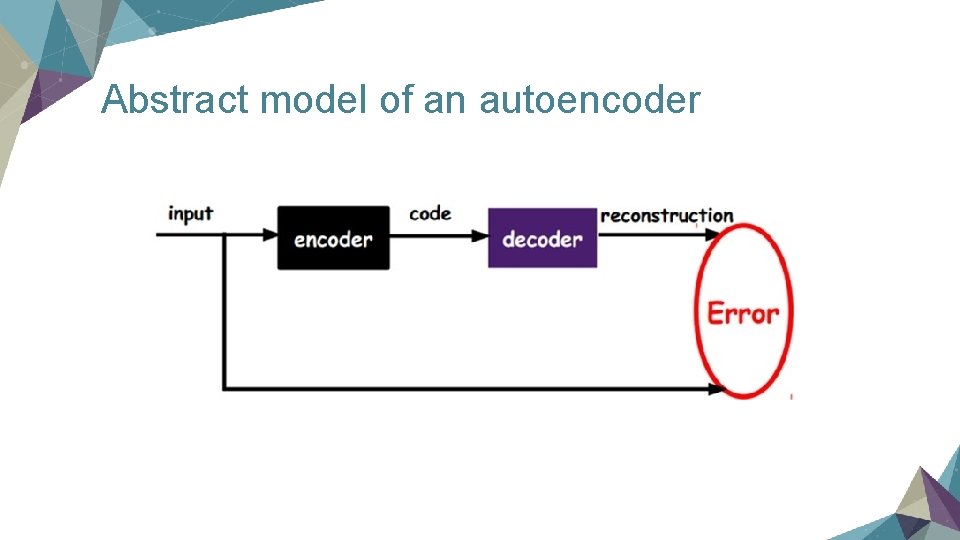
Abstract model of an autoencoder

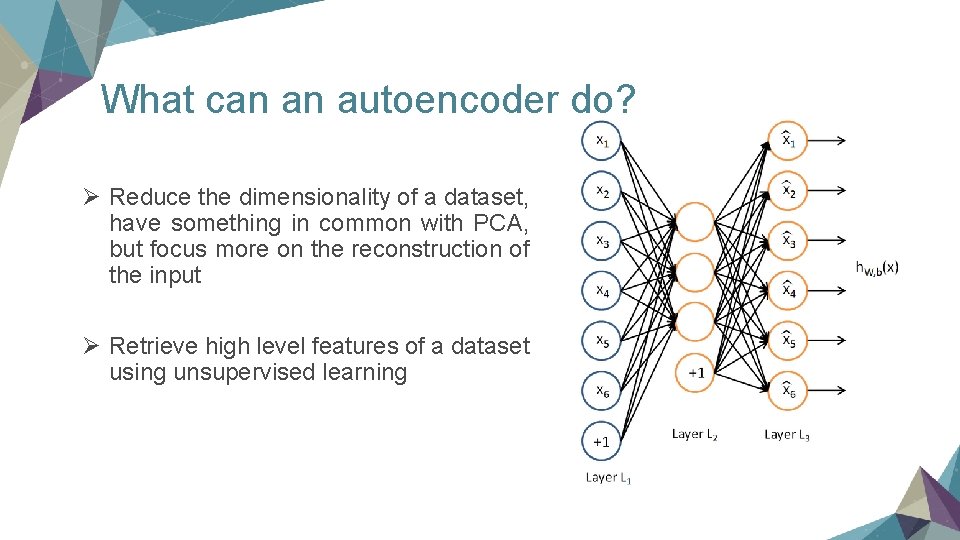
What can an autoencoder do? Ø Reduce the dimensionality of a dataset, have something in common with PCA, but focus more on the reconstruction of the input Ø Retrieve high level features of a dataset using unsupervised learning

Different types of autoencoder Ø Sparse Autoencoder Ø Stacked Autoencoder Ø Denoising Autoencoder

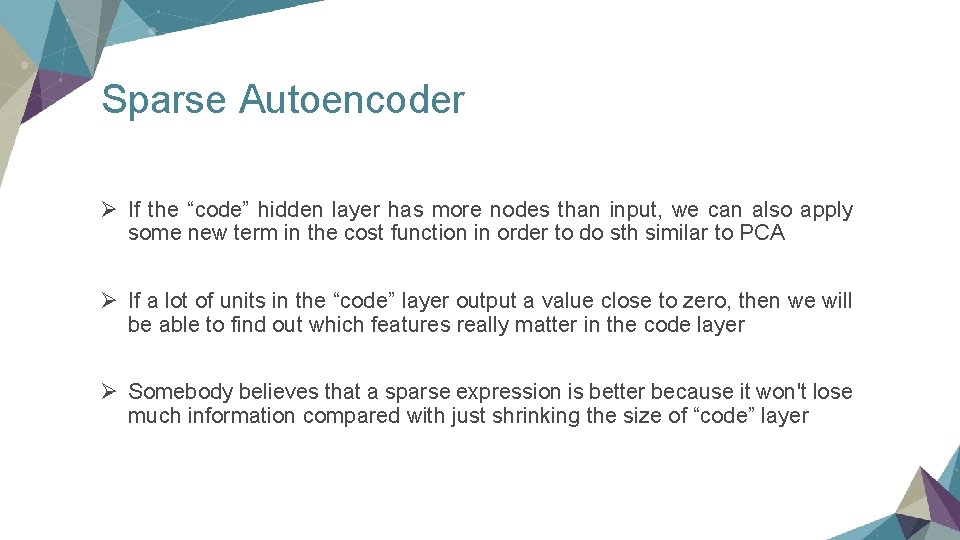
Sparse Autoencoder Ø If the “code” hidden layer has more nodes than input, we can also apply some new term in the cost function in order to do sth similar to PCA Ø If a lot of units in the “code” layer output a value close to zero, then we will be able to find out which features really matter in the code layer Ø Somebody believes that a sparse expression is better because it won't lose much information compared with just shrinking the size of “code” layer

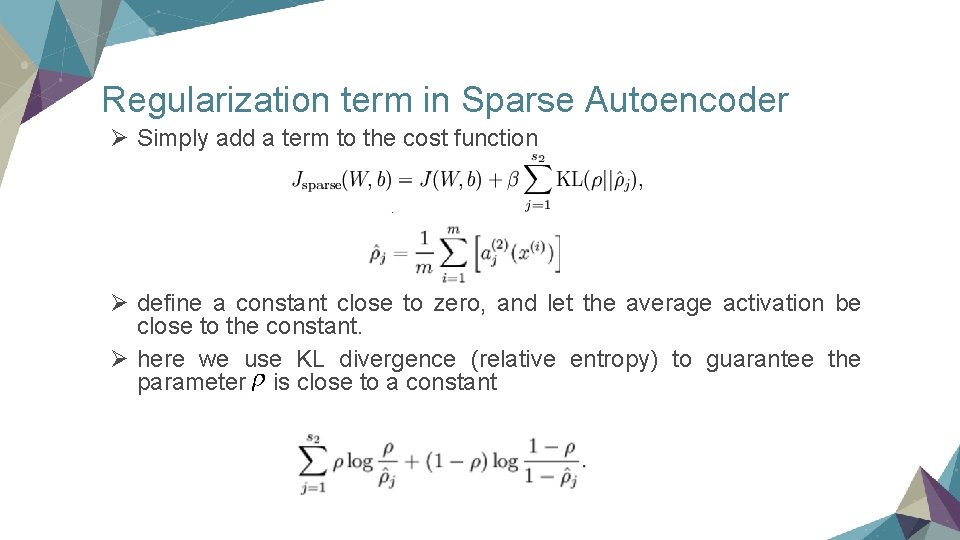
Regularization term in Sparse Autoencoder Ø Simply add a term to the cost function Ø define a constant close to zero, and let the average activation be close to the constant. Ø here we use KL divergence (relative entropy) to guarantee the parameter is close to a constant

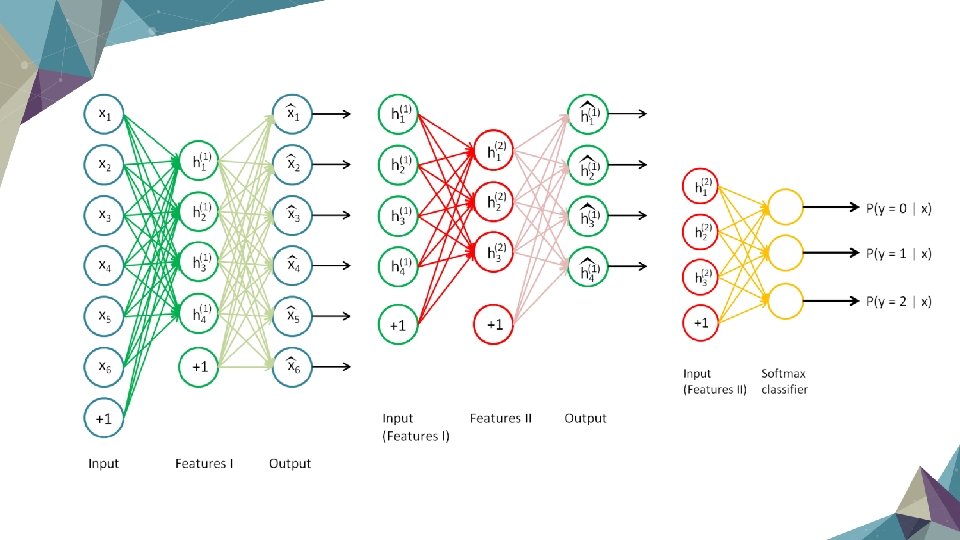
Stacked Autoencoders Ø Sometimes an autoencoder with single hidden layer is not strong enough to genertate good result Ø Make autoencoder like “stack” to get a deep network


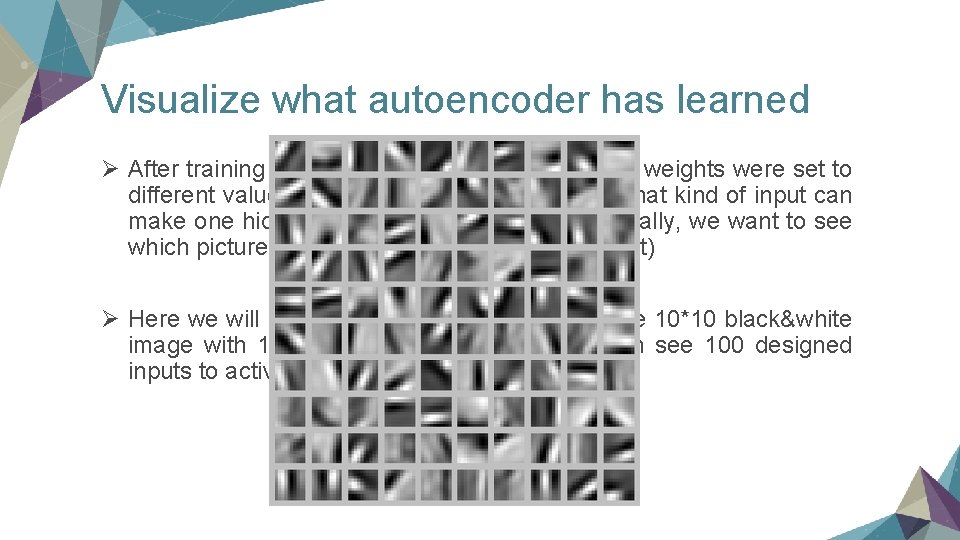
Visualize what autoencoder has learned Ø After training by a lot of data without labels, the weights were set to different values. In this case, we want to see what kind of input can make one hidden unit activate most. (Less formally, we want to see which picture the specific hidden unit wants most) Ø Here we will see a example: a model's input are 10*10 black&white image with 100 hidden units. (So here we can see 100 designed inputs to activate one of the 100 units most)

Some notes on autoencoders Ø “auto” here stands for unsupervised learning, it can retrieve useful features even without an output y of input features. Ø Autoencoder is often trained by deep learning methods, like greedy layer-wise training or pre-training with restricted Boltzman machine. Ø If you just train a deep autoencoder using normal back propagation just as mentioned before, it will suffer from gradient vanish a lot and will not be trained efficiently.

Part 5 Summary

Summary Ø Neural Network has always been a powerful model in machine learning. In bioinformatics we can use some simple network to help solving some regression problem without using deep learning methods. Ø Autoencoder as a tool of unsupervised learning can help us figure out some inner features of gene data. Also helpful in dimensionality reduction. Ø Beats other methods for its non-linear transformation strategy

Deep learning in practice Ø Powerful tool but hard to use in practice Ø You don't know the best number of nodes in each layer, and the number of layers as well. That's the main challenge Ø Tuning process is very time consuming, e. g. learning rate, dropout rate, momentum factor etc. You will never know what's the best. Ø Neural Network is not equal to deep learning, simple back propagation will fail in many cases, how to pre-train the model is also hard.

Reference 1. Stanford Machine Learning Online Course: https: //www. coursera. org/learn/machine-learning 2. UFLDL Tutorial: http: //ufldl. stanford. edu/wiki/index. php/UFLDL_Tutorial 3. I Sutskever,J Martens,G Dahl,G Hinton: On the importance of initialization and momentum in deep learning, International Conference on Machine Learning, 2013 4. G. E. Hinton∗, N. Srivastava, A. Krizhevsky, I. Sutskever and R. R. Salakhutdinov: Improving neural networks by preventing co-adaptation of feature detectors, 《Computer Science》, 2012, 3(4): pages. 212 -223 5. N Srivastava,G Hinton,A Krizhevsky,I Sutskever,R Salakhutdinov: Dropout: a simple way to prevent neural networks from overfitting, 《Journal of Machine Learning Research》, 2014, 15(1): 1929 -1958 6. Y Chen,Y Li,R Narayan,A Subramanian,X Xie: Gene expression inference with deep learning, 《 Bioinformatics》, 2016

7. G. E. Hinton* and R. R. Salakhutdinov: Reducing the Dimensionality of Data with Neural Networks, 《Science》, 2006, 313(5786): 504 -507 8. Logistic Regression & information theory: John Mount: The equivalence of logistic regression and maximum entropy models

Thanks for your attention! Yimin Hu Peking University Computer Science undergraduate Email: huyimin@pku. edu. cn