A Sublinear Algorithm For Weakly Approximating Edit Distance

A Sublinear Algorithm For Weakly Approximating Edit Distance Batu, Ergun, Killian, Magen, Raskhodnikova, Rubinfeld, Sami Presentation by Itai Dinur

Edit Distance (Levenshtein distance) l Let A, B be two strings over a fixed alphabet Σ. The edit distance D(A, B) between A and B is defined as the minimum number of character insertions, deletions, and substitutions that transform A into B, or vice versa.

Applications l l l Bioinformatics Text processing Web search

Algorithms l l Wagner and Fischer gave a dynamic programming algorithm that runs in time O(n 2) Masek and Paterson gave an improved algorithm that runs in time O(n 2/logn)

The Edit Distance Testing Problem l On input A, B and parameters 0<α<1, C>1: l l l Note that the output is unrestricted for nα<D(A, B)≤n/C l l If D(A, B)≤nα, output CLOSE with probability at least 2/3 If D(A, B)>n/C, output FAR with probability at least 2/3 E. g. cannot distinguish between n 0. 1 and n 0. 9 The algorithm presented for the problem runs in time Õ(nmax{α/2, 2α-1})

Motivation l In some applications, given many pairs of strings, one is interested in computing the edit distance only for close strings l For string pairs where the edit distance is above a certain threshold, the actual value of the distance is irrelevant

Lower Bound l Any probabilistic algorithm for the edit distance test problem requires Ω(nα/2) queries l The algorithm presented for the problem runs in time Õ(nmax{α/2, 2α-1}), which is close to optimal for α≤ 2/3

Other Approximations l There are several papers that give better approximation results, but none run in sublinear time l Andoni and Onak give an algorithm that computes the edit distance between two strings up to a factor of in n 1+o(1) time

Algorithm Overview l A recursive divide and conquer algorithm l l l B is broken into substrings which are recursively matched against A The matches are pieced together to form a matching for A It is too expensive to match all the substrings l A small number of them are sampled and matched, relying on statistical properties of the matchings
![Approximate Matching l Definition 1: An interval I = B[s…e] has a (t, E) Approximate Matching l Definition 1: An interval I = B[s…e] has a (t, E)](http://slidetodoc.com/presentation_image/3de7a878c9d264f6fc1dc38485e9e3a4/image-10.jpg)
Approximate Matching l Definition 1: An interval I = B[s…e] has a (t, E) -(approximate) matching with respect to A if for some interval A[s’…e’], s’=s+t and D(A[s’…e’], I)≤E A abcd 1234 efgh 5678 B cd 02 I has a (2, 1)-(approximate) matching with respect to A

Coordinated Matching l Definition 2: Let I = (I 1, …Ik) be a collection of intervals. We say that I has a (t, σ, E, D)coordinated matching with A if for all but D of the intervals Ii I, Ii has a (ti, E)-matching with A, where |t-ti|≤σ A abcd 1234 efgh 5678 B cd 0236 gjfkl 5 I has a (1, 1, 2, 1)-coordinated matching with A
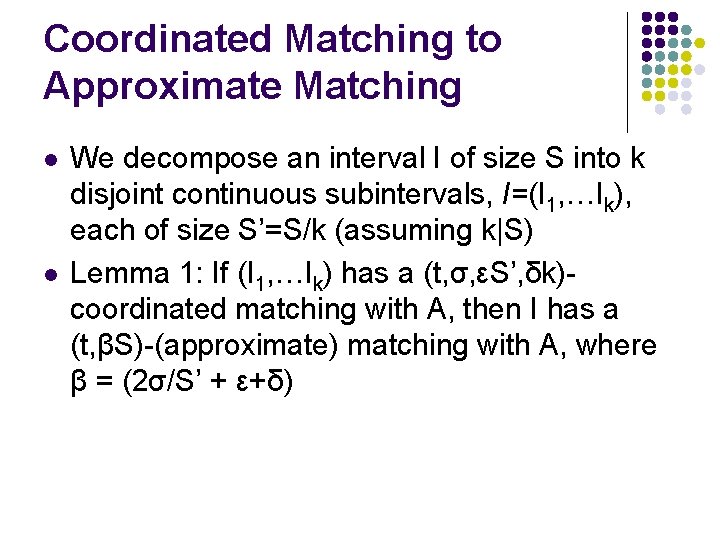
Coordinated Matching to Approximate Matching l l We decompose an interval I of size S into k disjoint continuous subintervals, I=(I 1, …Ik), each of size S’=S/k (assuming k|S) Lemma 1: If (I 1, …Ik) has a (t, σ, εS’, δk)coordinated matching with A, then I has a (t, βS)-(approximate) matching with A, where β = (2σ/S’ + ε+δ)

Approximate Matching to Coordinated Matching l l Lemma 2: Let c>1 and S>c. E. If I has a (t, E)matching with A then I=(I 1, …Ik) has (t, E, c. E/k, k/c)-coordinated matching with A Lemma 3: If I has a (t, E)-matching with A, and k≥E, then I=(I 1, …Ik) has (t, E, 0, E)coordinated matching with A

To match A and B l Decompose B into a set of continuous disjoint intervals I l l Lemma 2 argues that a match for A and B gives a coordinated matching for A and I Use a subroutine (COORD-MATCHES) to find coordinated matches for I l Lemma 1 infers the existence of good matches for B from coordinated matches for I

COORD-MATCHES l COORD-MATCHES(A, I, σ, E, D, ε, c) l l Let d be a constant, l=dlog(n). Choose samples i 1, …, il uniformly and independently from [1, …, k] For each chosen sample ij compute Tj=MATCHES(A, ij, E) Let Δ=(D/k+ε/2)l Return the set T, where t T iff Tj∩[t-σ…t+σ]=Ø for at most Δ sets Tj

Sampling Lemma l Lemma 4: Suppose that a random element of a set S of size n has a property Z with probability p. For any positive ε and c, there exists d such that for dlog(n) random samples from S the fraction p’ of these samples with property Z satisfies p-ε/2≤p’≤p+ε/2 with probability 1 -1/nc

COORD-MATCHES l Lemma 5: With probability 1 -1/nc-1 over the random coins of COORD-MATCHES, the output T of COORD-MATCHES(A, I, σ, E, D, ε, c) has the following properties: l l If I has a (t, σ, E, D)-coordinated matching then t T If t T then I has a (t, σ, E, D+εk)-coordinated matching

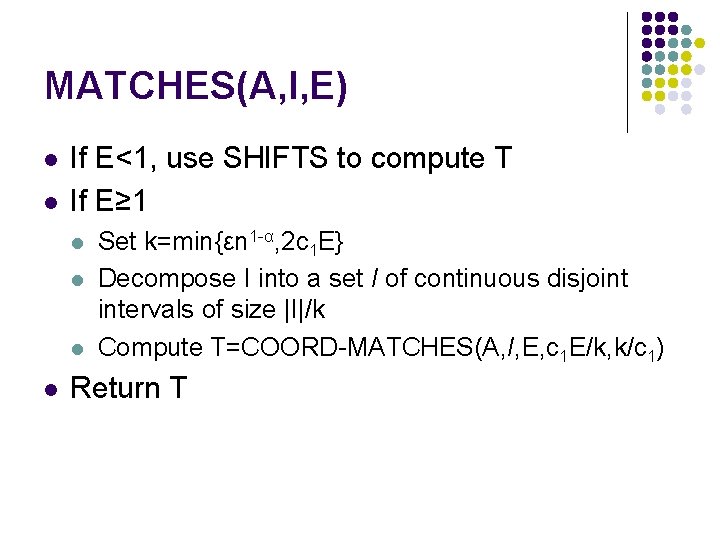
MATCHES(A, I, E) l l If E≥ 1, use a recursive call to COORDMATCHES If E<1 (i. e E=0), then A must contain the interval I unchanged. The set of t values is computed directly using the algorithm SHIFTS
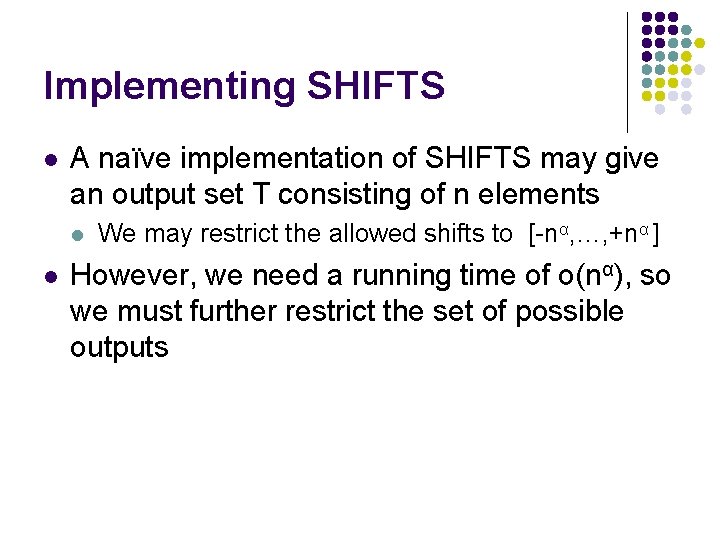
Implementing SHIFTS l A naïve implementation of SHIFTS may give an output set T consisting of n elements l l We may restrict the allowed shifts to [-nα, …, +nα ] However, we need a running time of o(nα), so we must further restrict the set of possible outputs

The Approximate Matching problem l Actually, we will solve the approximate matching problem: Given a block I=B[s…e] of length b=e-s+1, and a constant c 2>1, find all indexes s’ such that A[s’…(s’+b-1)] matches I, in a sense that the two substrings have Hamming-distance at most b/c 2 l Note that if D(A, B)<nα, it is enough to consider s’ in the interval [s-nα, s+nα]

The Approximate Matching problem l Naively, we can randomly sample O(log(n)) indexes i to determine (with high probability) if a substring of A[(t+1)…(t+b)] matches I, for a given t, and try all 2 nα possible shifts l Requires Ω(nα) queries to A
![The Ruler Procedure l l l We can compare pairs of characters A[i], I[j] The Ruler Procedure l l l We can compare pairs of characters A[i], I[j]](http://slidetodoc.com/presentation_image/3de7a878c9d264f6fc1dc38485e9e3a4/image-22.jpg)
The Ruler Procedure l l l We can compare pairs of characters A[i], I[j] such that a pair is compared for every i-j from 0 to u=2 nα with √u queries to each string given that b>√u In A character positions divisible by √u are queried A[√u, 2√u, …u]. In I, √u consecutive positions are queried I[1…√u] Define cen=ët/√uû+1 mil=t(mod√u), then for i=cen√u, j=√u-mil i-j=t
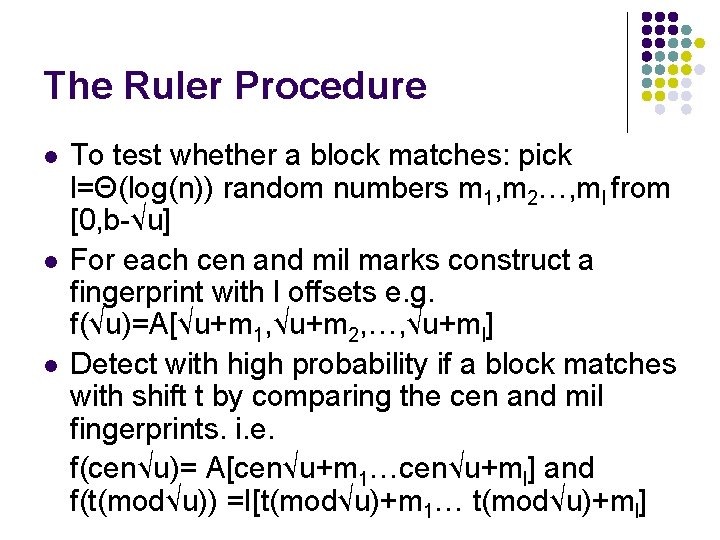
The Ruler Procedure l l l To test whether a block matches: pick l=Θ(log(n)) random numbers m 1, m 2…, ml from [0, b-√u] For each cen and mil marks construct a fingerprint with l offsets e. g. f(√u)=A[√u+m 1, √u+m 2, …, √u+ml] Detect with high probability if a block matches with shift t by comparing the cen and mil fingerprints. i. e. f(cen√u)= A[cen√u+m 1…cen√u+ml] and f(t(mod√u)) =I[t(mod√u)+m 1… t(mod√u)+ml]

The Ruler Procedure l l If b≤√u we have only O(b) mil marks and Ω(u/b) cen marks We can find all matching shifts by using O(max{√u, u/b}log(n)) queries
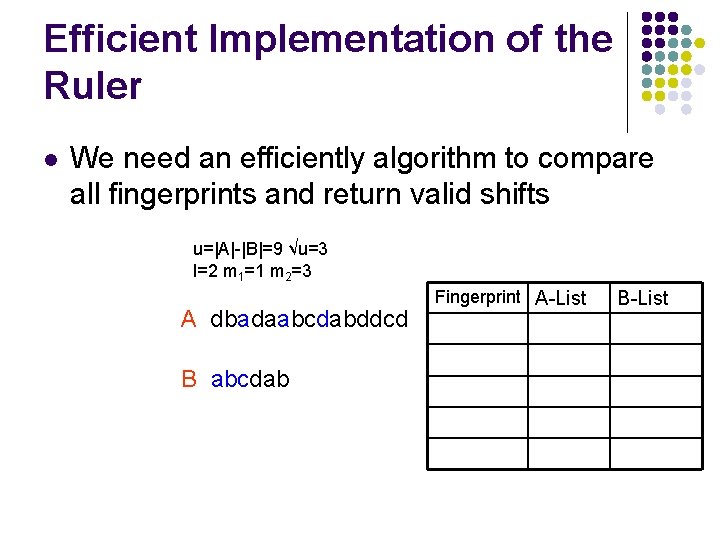
Efficient Implementation of the Ruler l We need an efficiently algorithm to compare all fingerprints and return valid shifts u=|A|-|B|=9 √u=3 l=2 m 1=1 m 2=3 A dbadaabcdabddcd B abcdab Fingerprint A-List B-List
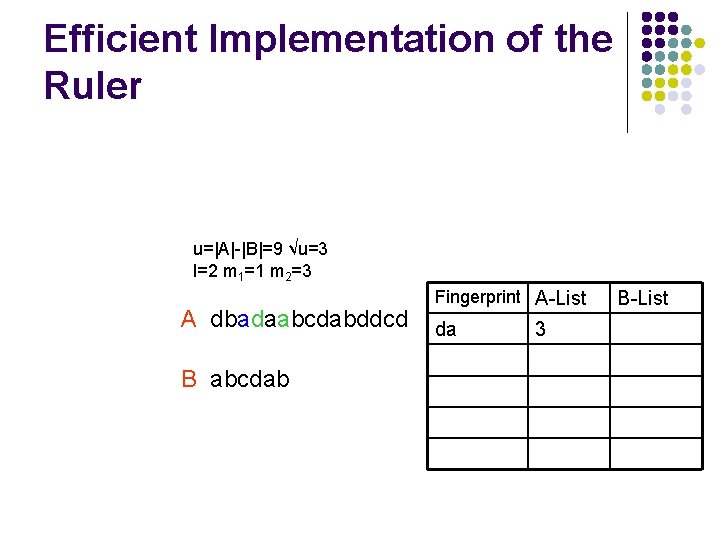
Efficient Implementation of the Ruler u=|A|-|B|=9 √u=3 l=2 m 1=1 m 2=3 A dbadaabcdabddcd B abcdab Fingerprint A-List da 3 B-List
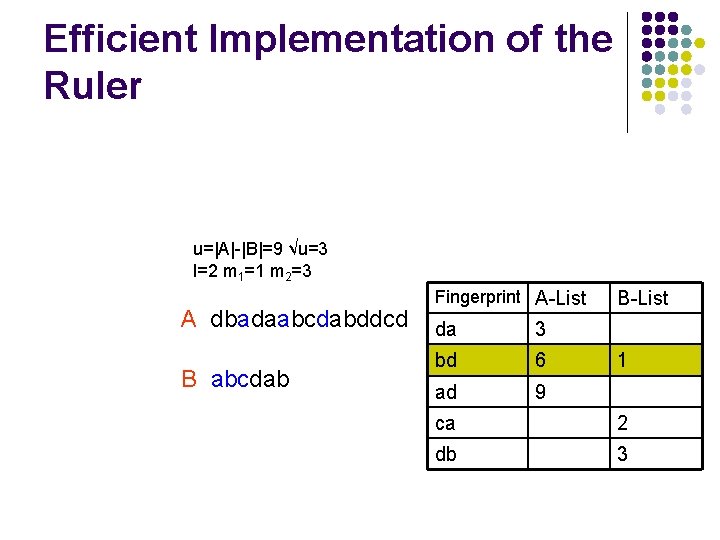
Efficient Implementation of the Ruler u=|A|-|B|=9 √u=3 l=2 m 1=1 m 2=3 A dbadaabcdabddcd B abcdab Fingerprint A-List da 3 bd 6 ad 9 B-List 1 ca 2 db 3

Quantizing the Ruler l l l The explicit list of all matching t can have Ω(u) values We round the values of t to multiples of some integer Q and return all quantized shifts The running time is O(max{√u, u/b, u/Q}log(n))

SHIFTS(A, I, Q) l l l Initialize the fingerprint data structure Pick l=Θ(log(n)) random numbers m 1, m 2…, ml Add all the fingerprints f(i) of A to the data structure, adding i to the A-list of f(i) Add all the fingerprints f(j) of I to the data structure, adding j to the B-list of f(j) Quantize all A-lists and B-lists For each fingerprint, output the list of quantized shifts (differences)

SHIFTS(A, I, Q) l Theorem 1: Procedure SHIFTS finds all quantized shifts of interval I in A, with high probability. It runs in time O(max{√u, u/b, u/Q}log(n)), where u=|A|-b
MATCHES(A, I, E) l l If E<1, use SHIFTS to compute T If E≥ 1 l l Set k=min{εn 1 -α, 2 c 1 E} Decompose I into a set I of continuous disjoint intervals of size |I|/k Compute T=COORD-MATCHES(A, I, E, c 1 E/k, k/c 1) Return T
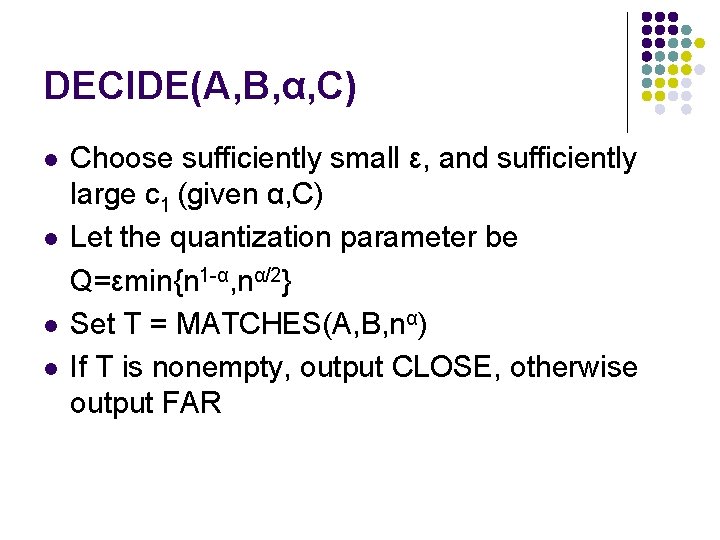
DECIDE(A, B, α, C) l l Choose sufficiently small ε, and sufficiently large c 1 (given α, C) Let the quantization parameter be Q=εmin{n 1 -α, nα/2} Set T = MATCHES(A, B, nα) If T is nonempty, output CLOSE, otherwise output FAR

DECIDE(A, B, α, C) l For any fixed α<1, we can choose constants ε and c 1 such that procedure DECIDE solves the edit distance testing problem with high probability

Running Time Analysis l l Note that when k=2 c 1 E, COORD-MATCHES is called with edit distance parameter c 1 E/k=1/2<1. I. e. next call to MATCHES will call SHIFTS and end the recursion Each level, The interval input to MATCHES goes down by a factor of k=Ω(n 1 -α), after r=α/(1 -α) levels the intervals are of length n/nr(1 -α)=O(n 1 -α), E=O(nα/nr(1 -α))=O(1) and SHIFT will be called next

Running Time Analysis α<1/2 l l l One level of recursion B is broken to intervals of size O(nα) dlog(n) calls to SHIFT with Q=εnα/2 Each call takes O(max{√u, u/b, u/Q}log(n)) = O(max{nα/2, 1, nα/2}log(n))=O(nα/2 log(n)) One merge taking O(nα/2 log(n)) Total running time O(nα/2 log 2(n))

Running Time Analysis 1/2<α<2/3 l l l Two levels of recursion At the last level, B is broken to intervals of size O(nα/2) log 2(n) calls to SHIFT with Q=εnα/2 Each call takes O(nα/2 log(n)) log(n) merges each taking O(nα/2 log(n)) Total running time O(nα/2 log 3(n))

Running Time Analysis α>2/3 l l l r>2 levels of recursion At the last level, B is broken to intervals of size O(n 1 -α) log. O(1)(n) calls to SHIFT with Q=εn 1 -α Note that n 1 -α<nα/2 Each call takes O(max{√u, u/b, u/Q}log(n)) = O((u/b)log(n))=O(n 2α-1 log(n)) Total running time Õ(n 2α-1 log(n))

Conclusion l l We saw an algorithm for the edit distance test problem that runs in time Õ(nmax{α/2, 2α-1}) Any probabilistic algorithm for the edit distance test problem requires Ω(nα/2) queries
- Slides: 38