A Robust Framework for Detecting Structural Variations February

A Robust Framework for Detecting Structural Variations February 6, 2008 Seunghak Lee 1, Elango Cheran 1, and Michael Brudno 1 1 University of Toronto, Canada 1

What are structural variations? (1) n 10^3 – 10^6 basepair variations in the genome n Insertion: a large consecutive fragment of DNA is inserted n Deletion: a large consecutive fragment of DNA is deleted n Inversion: a large consecutive fragment of DNA is inversed n Translocation: a large consecutive fragment of DNA is moved from one chromosome to another. n Copy number variations 2

What are structural variations? (2) Various examples of structural variations 3

Outline n Introduction Type of Structural Variations ¨ Sequencing Approaches to Detect Structural Variations ¨ Motivation & Research Objectives ¨ n Probabilistic Framework for Detecting Structural Variations Probabilistic Framework ¨ Flow of our Framework ¨ Hierarchical Clustering of Matepairs (2 nd phase) ¨ Choosing a Unique Mapped Location for Each Matepair (3 nd phase) ¨ n Experiments Comparison with Three Previous research ¨ DMBT 1 Gene for Deletion ¨ Centromere and Translocations ¨ n Conclusions 4
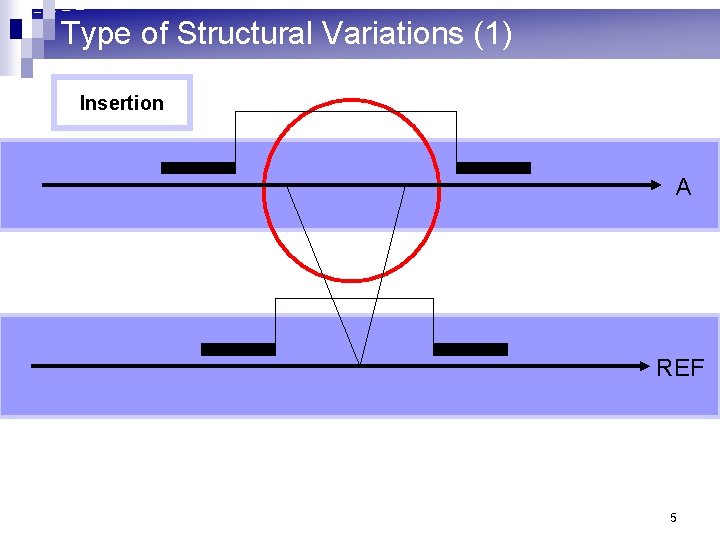
Type of Structural Variations (1) Insertion A REF 5
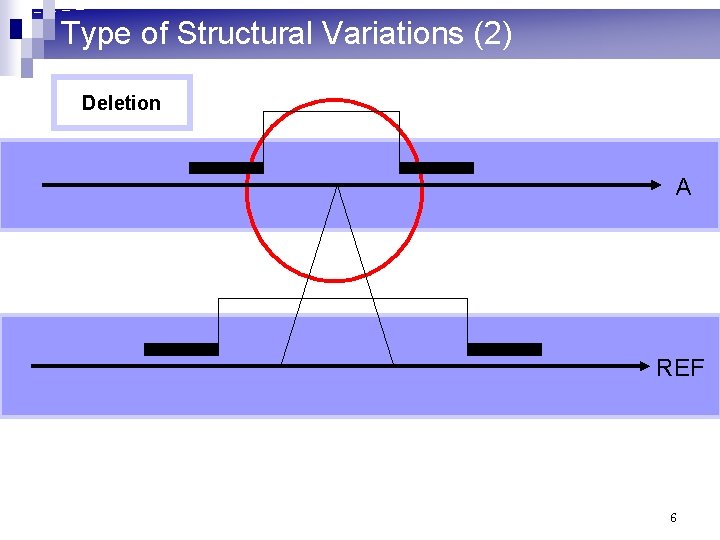
Type of Structural Variations (2) Deletion A REF 6
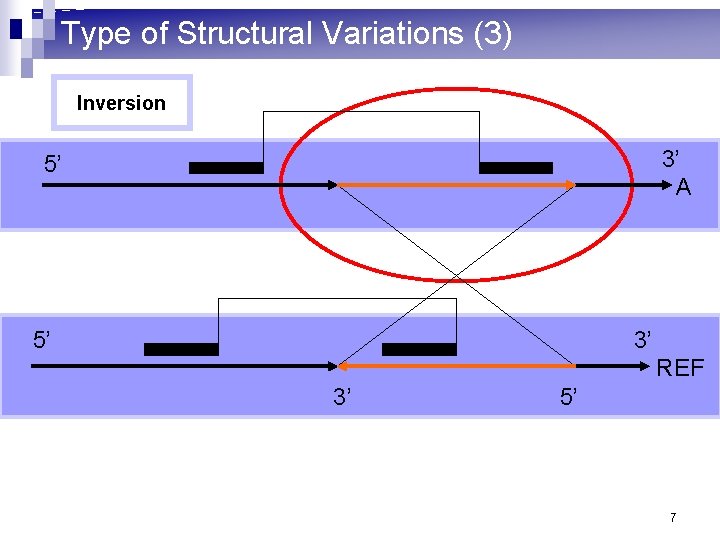
Type of Structural Variations (3) Inversion 3’ A 5’ 5’ 3’ REF 3’ 5’ 7
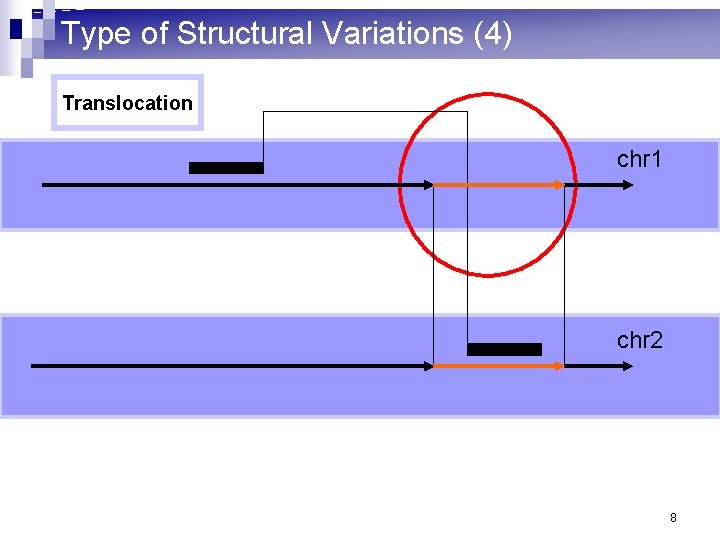
Type of Structural Variations (4) Translocation chr 1 chr 2 8
![Sequencing Approaches 1. “Fine-scale structural variation of the human genome” [Tuzun et al, 2005] Sequencing Approaches 1. “Fine-scale structural variation of the human genome” [Tuzun et al, 2005]](http://slidetodoc.com/presentation_image_h2/35c23a3b021e9f25e9b86e0e6bed0ae6/image-9.jpg)
Sequencing Approaches 1. “Fine-scale structural variation of the human genome” [Tuzun et al, 2005] • Mapping matepairs onto the reference genome • Insertion and deletion: inconsistent mapped distance • Inversion: the same orientation of both reads 2. “Paired-End mappings Reveals Extensive Structural Variation in the Human Genome” [Korbel et al, 2007] • Proposed high-throughput and massive paired end mapping technique • Detailed types of structural variations 9
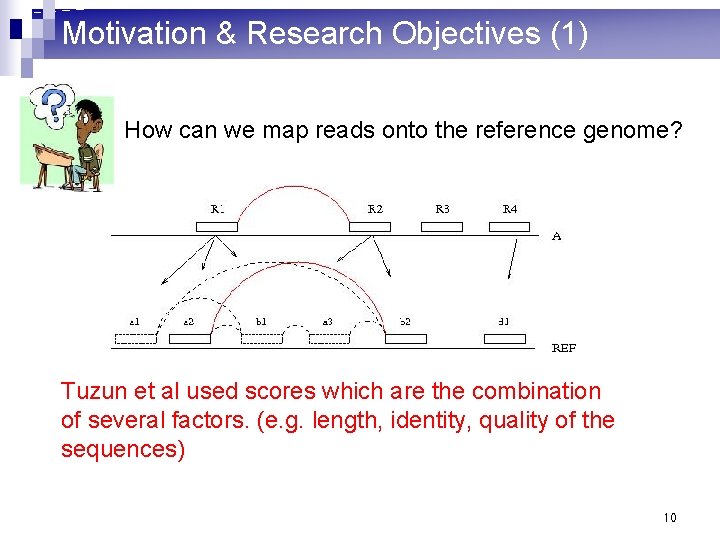
Motivation & Research Objectives (1) How can we map reads onto the reference genome? Tuzun et al used scores which are the combination of several factors. (e. g. length, identity, quality of the sequences) 10

Motivation & Research Objectives (2) n Sequencing method is effective to detect structural variants. ¨ n However, there are multiple mappings for each read ¨ n Proven by Tuzun et al, Korbel et al Previous research used a priori mapped locations. Why don’t we develop a probabilistic model without such assumptions? ¨ Hopefully, it can be applied to short reads from NGS machines. 11
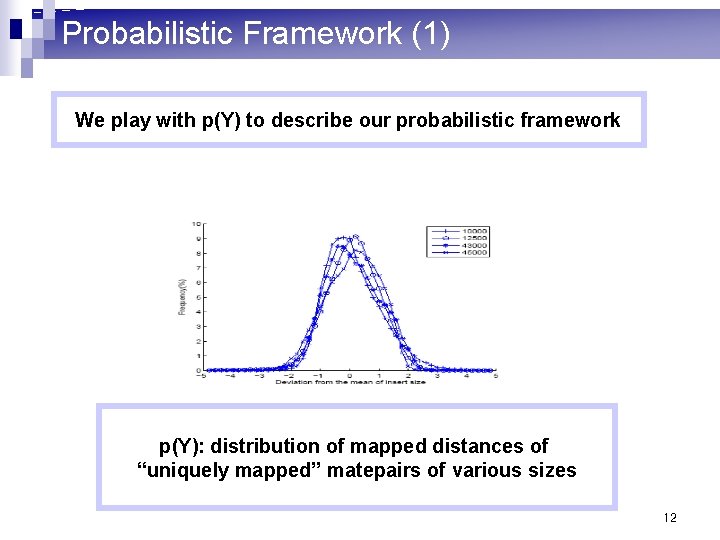
Probabilistic Framework (1) We play with p(Y) to describe our probabilistic framework p(Y): distribution of mapped distances of “uniquely mapped” matepairs of various sizes 12
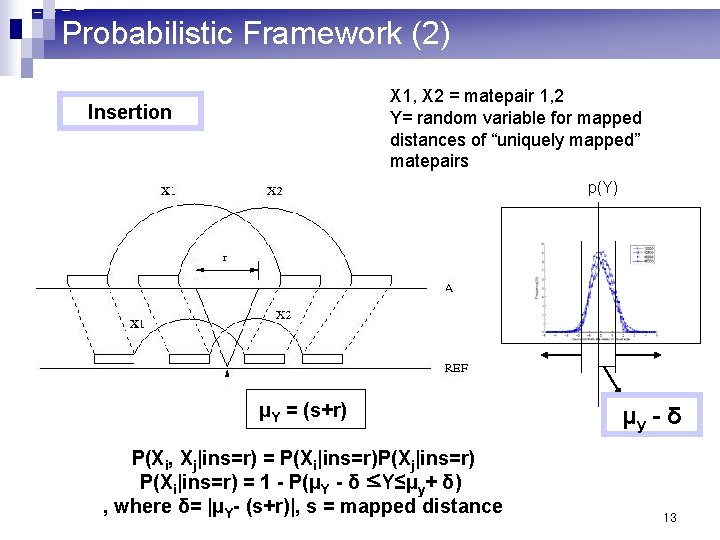
Probabilistic Framework (2) X 1, X 2 = matepair 1, 2 Y= random variable for mapped distances of “uniquely mapped” matepairs Insertion p(Y) μY = (s+r) P(Xi, Xj|ins=r) = P(Xi|ins=r)P(Xj|ins=r) P(Xi|ins=r) = 1 - P(μY - δ ≤Y≤μy+ δ) , where δ= |μY- (s+r)|, s = mapped distance μy - δ 13
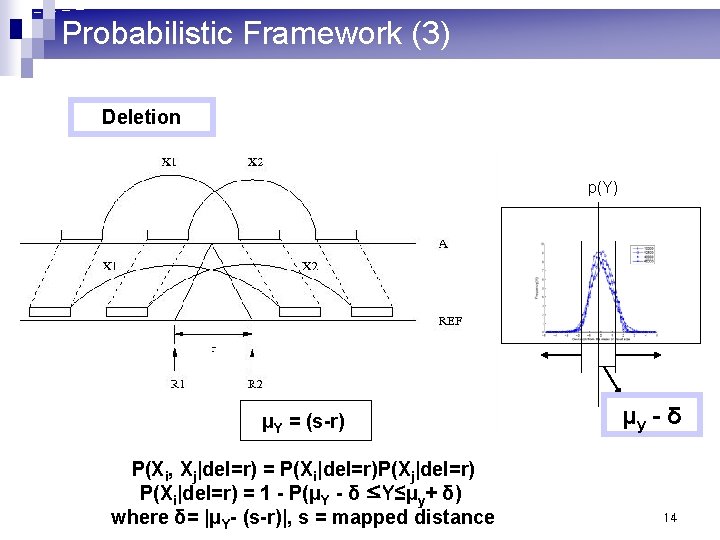
Probabilistic Framework (3) Deletion p(Y) μY = (s-r) P(Xi, Xj|del=r) = P(Xi|del=r)P(Xj|del=r) P(Xi|del=r) = 1 - P(μY - δ ≤Y≤μy+ δ) where δ= |μY- (s-r)|, s = mapped distance μy - δ 14
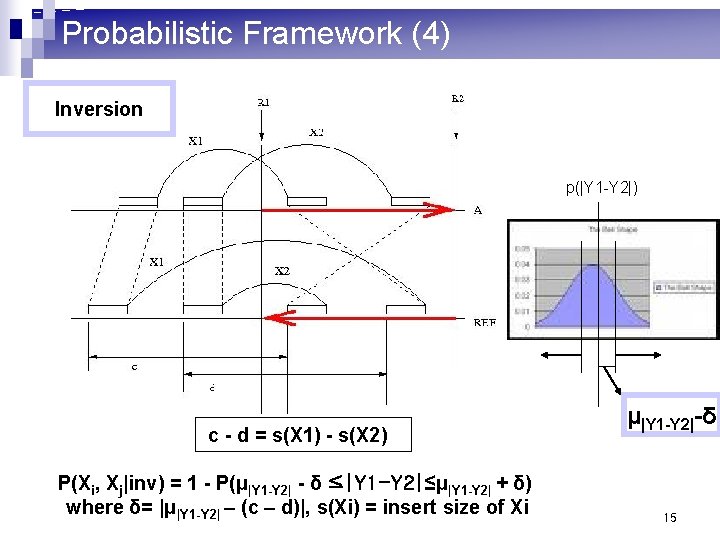
Probabilistic Framework (4) Inversion p(|Y 1 -Y 2|) c - d = s(X 1) - s(X 2) P(Xi, Xj|inv) = 1 - P(μ|Y 1 -Y 2| - δ ≤|Y 1 -Y 2|≤μ|Y 1 -Y 2| + δ) where δ= |μ|Y 1 -Y 2| – (c – d)|, s(Xi) = insert size of Xi μ|Y 1 -Y 2|-δ 15
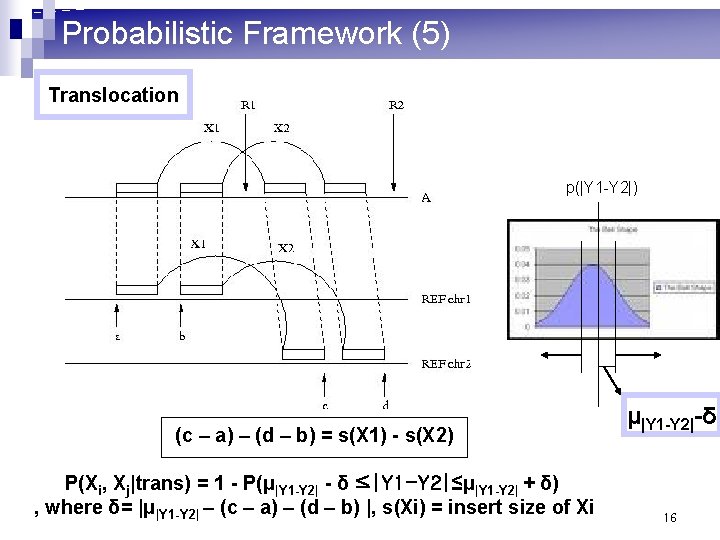
Probabilistic Framework (5) Translocation p(|Y 1 -Y 2|) (c – a) – (d – b) = s(X 1) - s(X 2) P(Xi, Xj|trans) = 1 - P(μ|Y 1 -Y 2| - δ ≤|Y 1 -Y 2|≤μ|Y 1 -Y 2| + δ) , where δ= |μ|Y 1 -Y 2| – (c – a) – (d – b) |, s(Xi) = insert size of Xi μ|Y 1 -Y 2|-δ 16
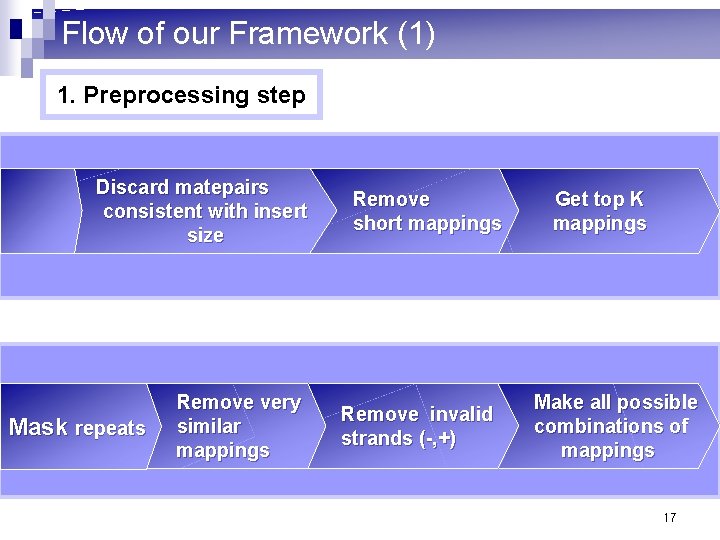
Flow of our Framework (1) 1. Preprocessing step Discard matepairs consistent with insert size Mask repeats Remove very similar mappings Remove short mappings Remove invalid strands (-, +) Get top K mappings Make all possible combinations of mappings 17
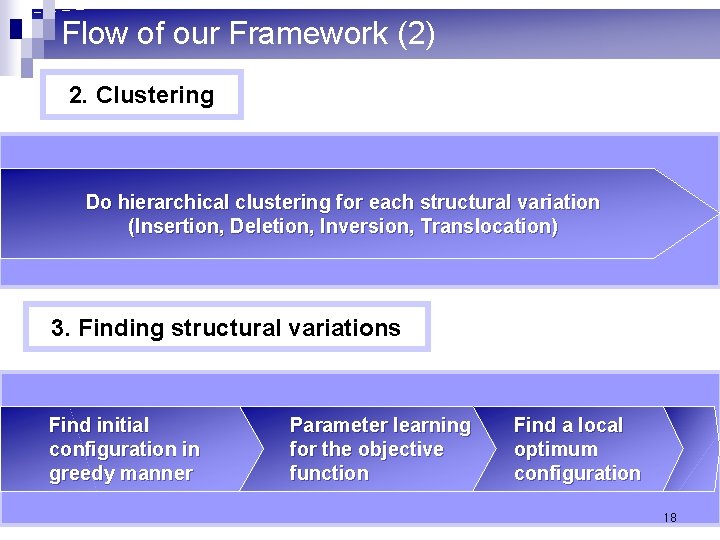
Flow of our Framework (2) 2. Clustering Do hierarchical clustering for each structural variation (Insertion, Deletion, Inversion, Translocation) 3. Finding structural variations Find initial configuration in greedy manner Parameter learning for the objective function Find a local optimum configuration 18
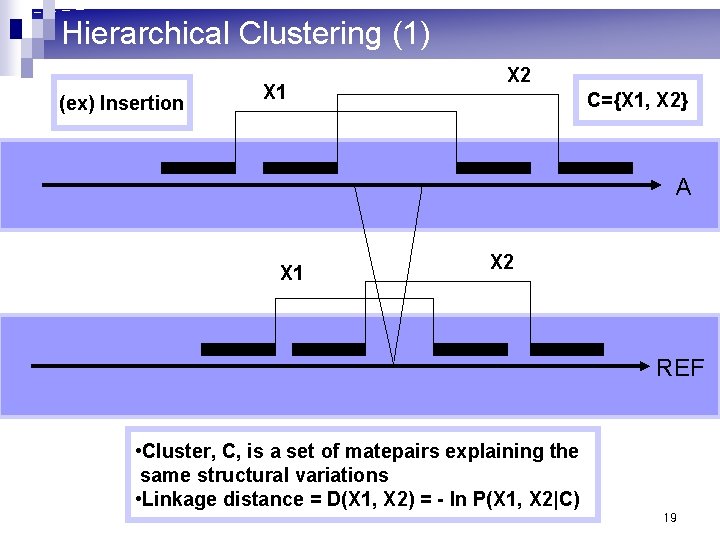
Hierarchical Clustering (1) (ex) Insertion X 1 X 2 C={X 1, X 2} A X 1 X 2 REF • Cluster, C, is a set of matepairs explaining the same structural variations • Linkage distance = D(X 1, X 2) = - ln P(X 1, X 2|C) 19
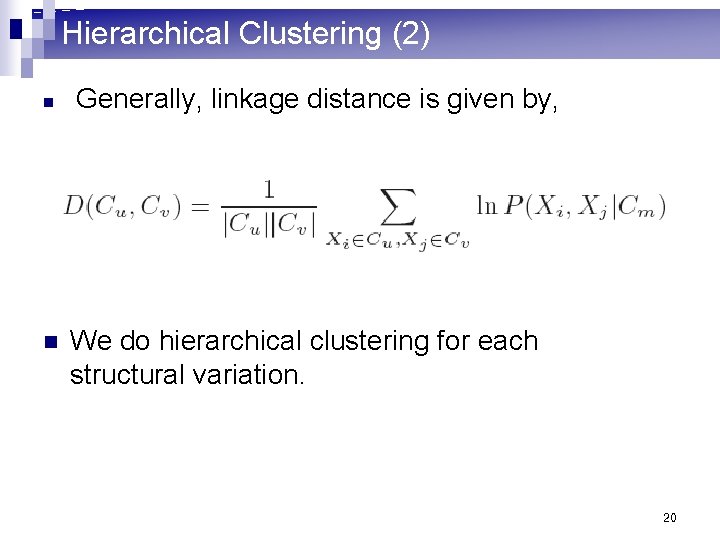
Hierarchical Clustering (2) n n Generally, linkage distance is given by, We do hierarchical clustering for each structural variation. 20
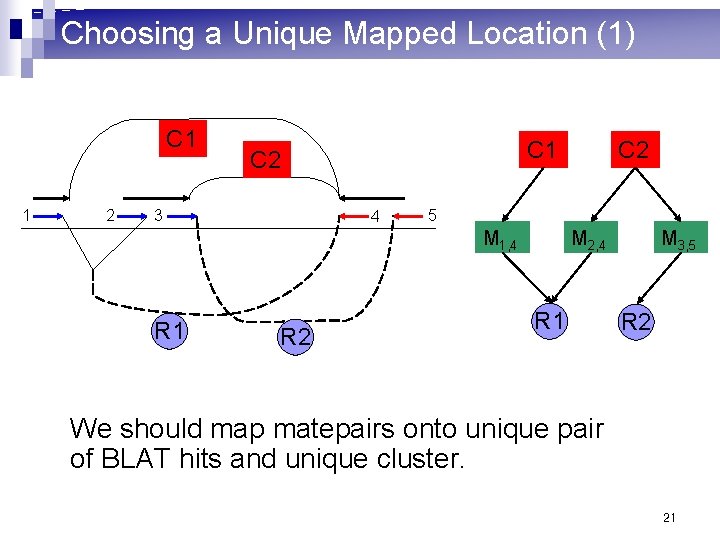
Choosing a Unique Mapped Location (1) C 1 1 2 C 1 C 2 3 4 5 M 1, 4 R 1 R 2 C 2 M 2, 4 R 1 M 3, 5 R 2 We should map matepairs onto unique pair of BLAT hits and unique cluster. 21
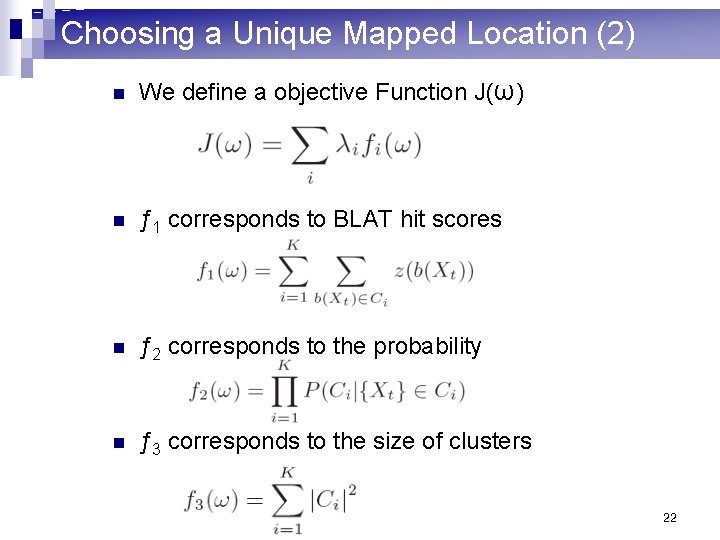
Choosing a Unique Mapped Location (2) n We define a objective Function J(ω) n ƒ 1 corresponds to BLAT hit scores n ƒ 2 corresponds to the probability n ƒ 3 corresponds to the size of clusters 22

Choosing a Unique Mapped Location (3) n Find the initial configuration greedily n Learn parameters for the objective function J(ω). ¨ We used hill climbing search to maximize the log likelihood of P(ω|λi) n Finally, find a configuration, locally maximizing J(ω) using hill climbing search 23
P-values n We assign p-values to give confidence to our clusters. n The probability that the cluster is generated by the reference genome not by structural variants ¨ Pval(Ck)=(E choose |Ck|) ∏ P(Xi|Cnull) where E = (Expected number of matepairs mapped to the location of the cluster) n P-values depend on the length of the cluster, thenumber of matepairs involved and probabilities. 24
Clustering Results We started with ~360, 000 matepair n ~90% were uniquely mapped n ~90% had a concordant position (mapped at ± 2 ) Through the clustering procedure above (FDR 0. 2) we found n n 82 Insertion clusters (53 had a uniquely mapped read) 175 Deletion clusters (135) 103 inversion clusters (24) 55 Translocation (cross-chromosome) cluster (all were required to have a uniquely mapped read) 25
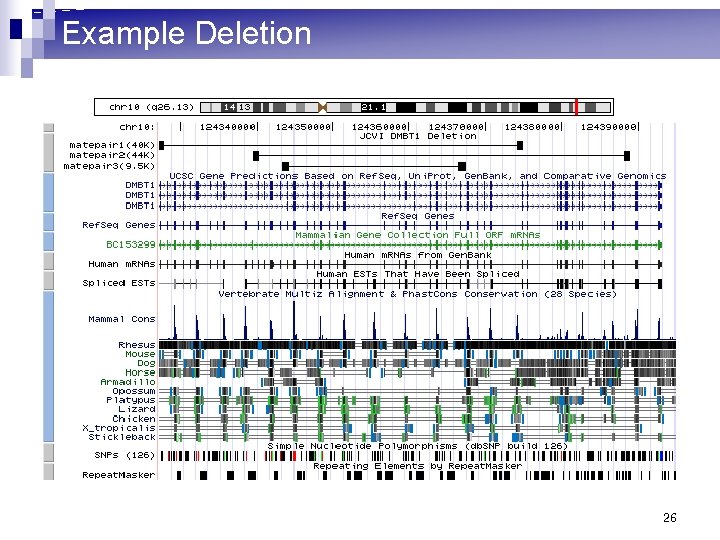
Example Deletion 26
Agreement with Previous Results Type Total Tuzun Levy Korbel DGV-All Insertion 82(53) 12(7)/139 6(5)/319 0(0)/34 24(13)/2216 Deletion 175(135) 21(17)/102 25(23)/344 45(36)/742 82(63)/4697 Inversion 103(24) 34(12)/56 N/A 42(8)/105 60(15)/164 All of the correlations (besides the zero) are significant (p-values < 0. 001) via Monte Carlo simulations We have compared The DMBT 1 deletion was also found in the Tuzun et al dataset (but not the Levy dataset). 27
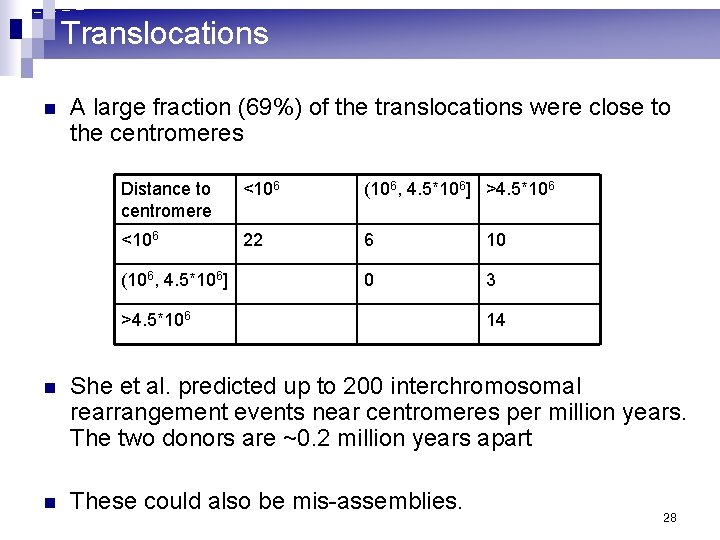
Translocations n A large fraction (69%) of the translocations were close to the centromeres Distance to centromere <106 (106, 4. 5*106] >4. 5*106 <106 22 6 10 0 3 (106, 4. 5*106] >4. 5*106 14 n She et al. predicted up to 200 interchromosomal rearrangement events near centromeres per million years. The two donors are ~0. 2 million years apart n These could also be mis-assemblies. 28
Conclusions n Introduced a novel framework for finding structural variants that does not rely on ab initio mapping of matepairs to genomic positions. n Introduced a probabilistic model for structural variants n Isolated 82 insertions, 175 deletions, and 103 inversions between the reference public human genome and the JCVI donor. n These results show statistically significant correlation with previous variation studies n Isolated 194 novel structural variants that do not overlap any event from the database of genomic variants (of these 121 have support from a uniquely mapped matepair) 29
- Slides: 29