A Radiation Hybrid Map of the Zebrafish Genome
“A Radiation Hybrid Map of the Zebrafish Genome” (Geisler et al. , 1999) D 145 Journal Club 1/16/2020 Daniel Benavides Matthieu Weber

Zebrafish as a Model Organism ● First used by George Streisinger at the University of Oregon in the 1980’s ● Zebrafish (Danio Rerio) is a desirable Model Organism ○ ○ Eggs developed outside of of the mother body: observable 70% genome shared with humans Cheaper than mice to maintain Lay as many as 300 eggs at a time (High N) Adult Zebrafish Photo: Max Planck Institute for Developmental Biology Zebrafish Embryo
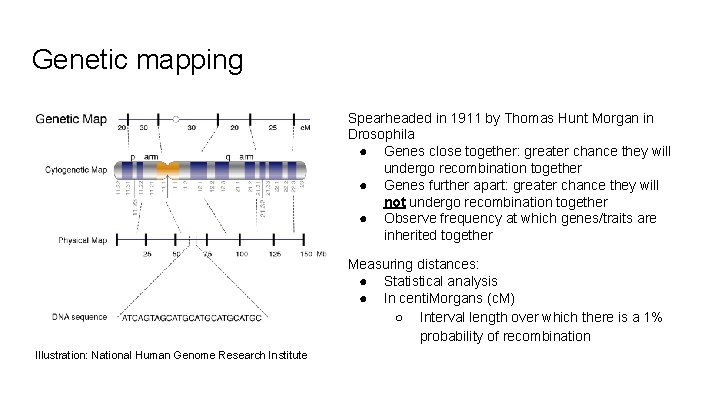
Genetic mapping Spearheaded in 1911 by Thomas Hunt Morgan in Drosophila ● Genes close together: greater chance they will undergo recombination together ● Genes further apart: greater chance they will not undergo recombination together ● Observe frequency at which genes/traits are inherited together Measuring distances: ● Statistical analysis ● In centi. Morgans (c. M) ○ Interval length over which there is a 1% probability of recombination Illustration: National Human Genome Research Institute

Previous Efforts: ● Prior studies have produced Radiation Hybrid maps of human, dog, mouse, and rat genomes Objectives: ● To create a high resolution map of the zebrafish whole genome using Radiation Hybridization Mapping ● validate/enhance previous genomic maps of the zebrafish
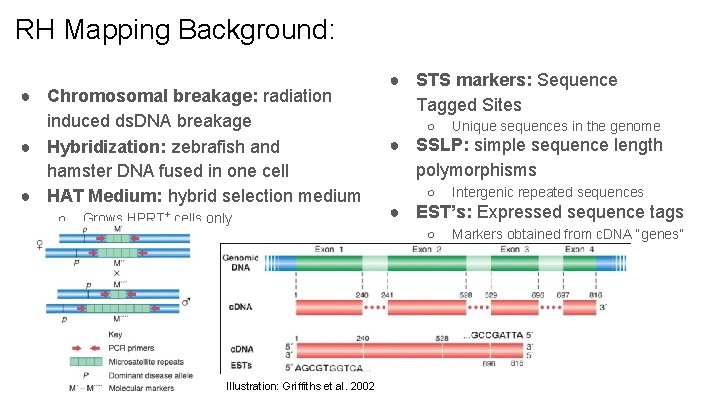
RH Mapping Background: ● Chromosomal breakage: radiation induced ds. DNA breakage ● Hybridization: zebrafish and hamster DNA fused in one cell ● HAT Medium: hybrid selection medium ○ Grows HPRT+ cells only ● STS markers: Sequence Tagged Sites ○ ● SSLP: simple sequence length polymorphisms ○ Intergenic repeated sequences ● EST’s: Expressed sequence tags ○ Illustration: Griffiths et al. 2002 Unique sequences in the genome Markers obtained from c. DNA “genes”
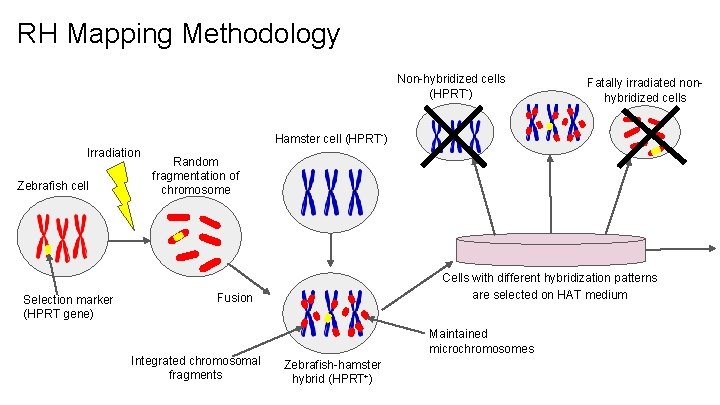
RH Mapping Methodology Non-hybridized cells (HPRT-) Fatally irradiated nonhybridized cells Hamster cell (HPRT-) Irradiation Zebrafish cell Selection marker (HPRT gene) Random fragmentation of chromosome Cells with different hybridization patterns are selected on HAT medium Fusion Integrated chromosomal fragments Maintained microchromosomes Zebrafish-hamster hybrid (HPRT+)
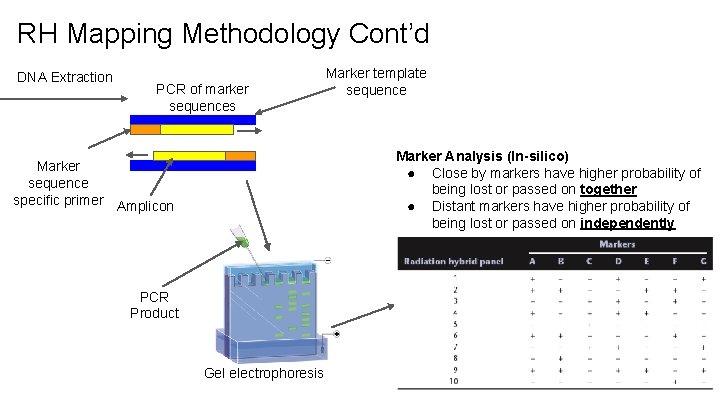
RH Mapping Methodology Cont’d DNA Extraction PCR of marker sequences Marker template sequence Marker Analysis (In-silico) ● Close by markers have higher probability of being lost or passed on together ● Distant markers have higher probability of being lost or passed on independently Marker sequence specific primer Amplicon PCR Product Gel electrophoresis
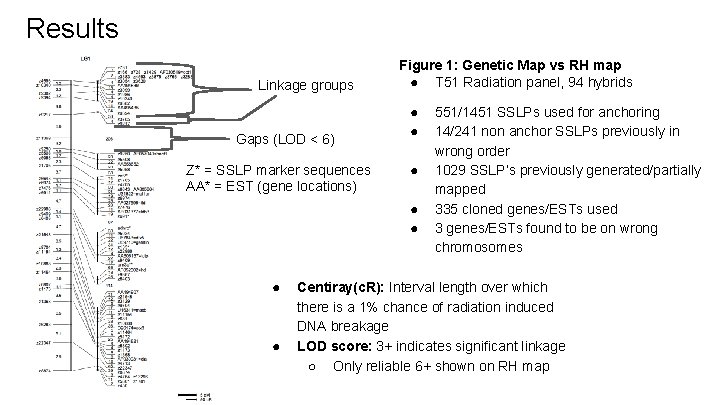
Results Linkage groups Gaps (LOD < 6) Z* = SSLP marker sequences AA* = EST (gene locations) Figure 1: Genetic Map vs RH map ● T 51 Radiation panel, 94 hybrids ● ● ● ● 551/1451 SSLPs used for anchoring 14/241 non anchor SSLPs previously in wrong order 1029 SSLP’s previously generated/partially mapped 335 cloned genes/ESTs used 3 genes/ESTs found to be on wrong chromosomes Centiray(c. R): Interval length over which there is a 1% chance of radiation induced DNA breakage LOD score: 3+ indicates significant linkage ○ Only reliable 6+ shown on RH map
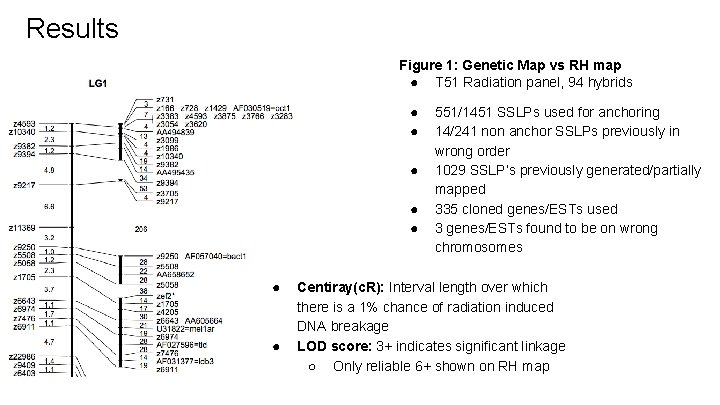
Results Figure 1: Genetic Map vs RH map ● T 51 Radiation panel, 94 hybrids ● ● ● ● 551/1451 SSLPs used for anchoring 14/241 non anchor SSLPs previously in wrong order 1029 SSLP’s previously generated/partially mapped 335 cloned genes/ESTs used 3 genes/ESTs found to be on wrong chromosomes Centiray(c. R): Interval length over which there is a 1% chance of radiation induced DNA breakage LOD score: 3+ indicates significant linkage ○ Only reliable 6+ shown on RH map
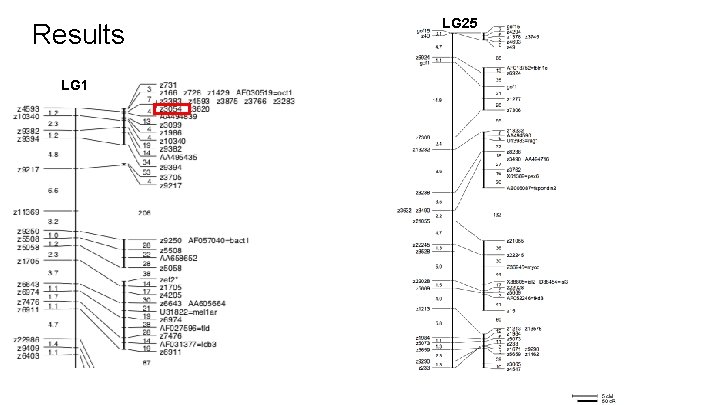
Results LG 1 c. M c. R LG 25
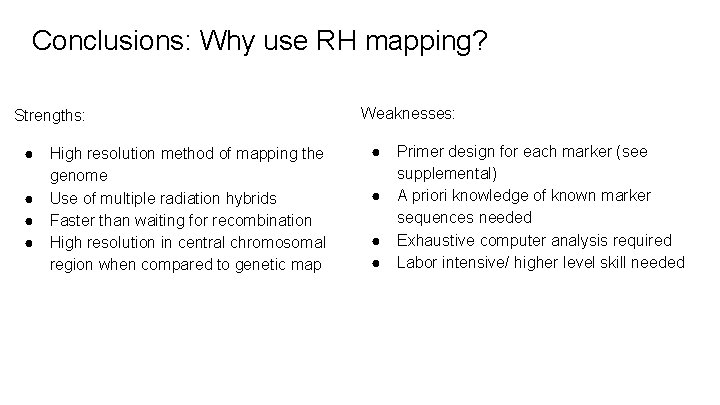
Conclusions: Why use RH mapping? Strengths: ● ● High resolution method of mapping the genome Use of multiple radiation hybrids Faster than waiting for recombination High resolution in central chromosomal region when compared to genetic map Weaknesses: ● ● Primer design for each marker (see supplemental) A priori knowledge of known marker sequences needed Exhaustive computer analysis required Labor intensive/ higher level skill needed

Main Takeaways ● ● ● First RH map of non-mammalian genome Provides a scaffold for mapping more ESTs Elucidates location of shared loci between zebrafish and humans Revealed discrepancies from previous genetic maps Facilitate the forward gene approach ○ Tracing mutant phenotypes back to their causative genes and their location in genome

Further Readings Gyapay, et al. , 1996: “RH map of the human genome” Cox, , et al. , 1990 “RH mapping in mammalian chromosomes” Mc. Carthy, et al. , 1997 “A First-Generation Whole Genome–Radiation Hybrid Map Spanning the Mouse Genome” Hukriede, et al. , 1999 "Radiation hybrid mapping of the zebrafish genome. " Boehnke et al. , 1991, "Statistical Methods for Multipoint Radiation Hybrid Mapping"

Works Cited (URL) http: //www. uvm. edu/~dstratto/bcor 101/0920. pdf https: //www. ndsu. edu/pubweb/~mcclean/plsc 431/linkage/linkag e 6. htm https: //www. sciencedirect. com/topics/biochemistry-geneticsand-molecular-biology/radiation-hybrid-mapping https: //pdfs. semanticscholar. org/304 c/a 89 f 276 e 4519 bb 2 b 23 e 9 b 786 d 0 f 21 b 6 f 7 f 3 f. pdf
- Slides: 14