A PARALLEL ALGORITHM FOR EXTRACTING TRANSCRIPTIONAL REGULATORY NETWORK
















- Slides: 30

A PARALLEL ALGORITHM FOR EXTRACTING TRANSCRIPTIONAL REGULATORY NETWORK MOTIFS Fu Rong Wu

OUTLINE Preliminary Previous Work Method Experimental Conclusion Result

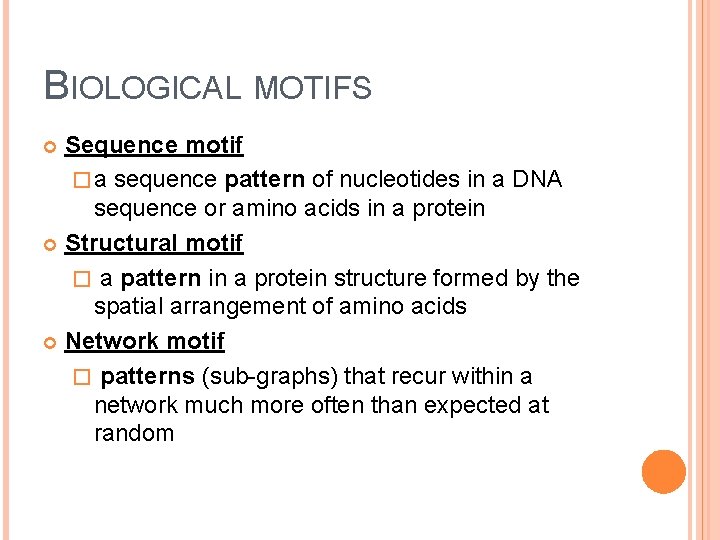
BIOLOGICAL MOTIFS Sequence motif � a sequence pattern of nucleotides in a DNA sequence or amino acids in a protein Structural motif � a pattern in a protein structure formed by the spatial arrangement of amino acids Network motif � patterns (sub-graphs) that recur within a network much more often than expected at random

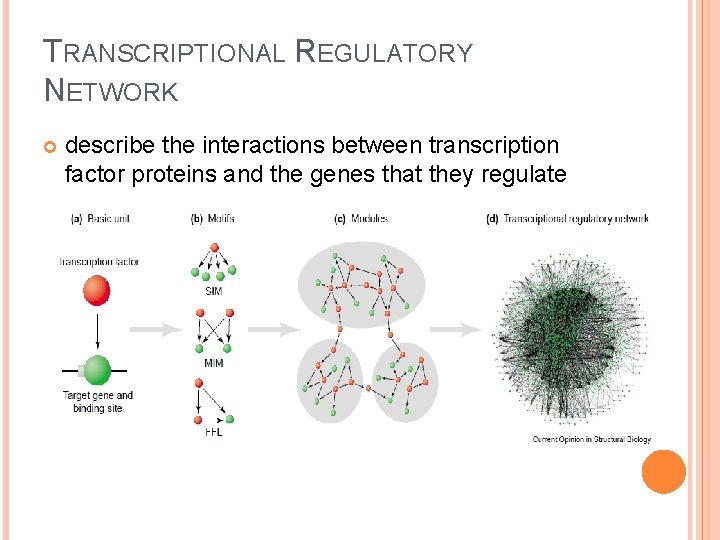
TRANSCRIPTIONAL REGULATORY NETWORK describe the interactions between transcription factor proteins and the genes that they regulate

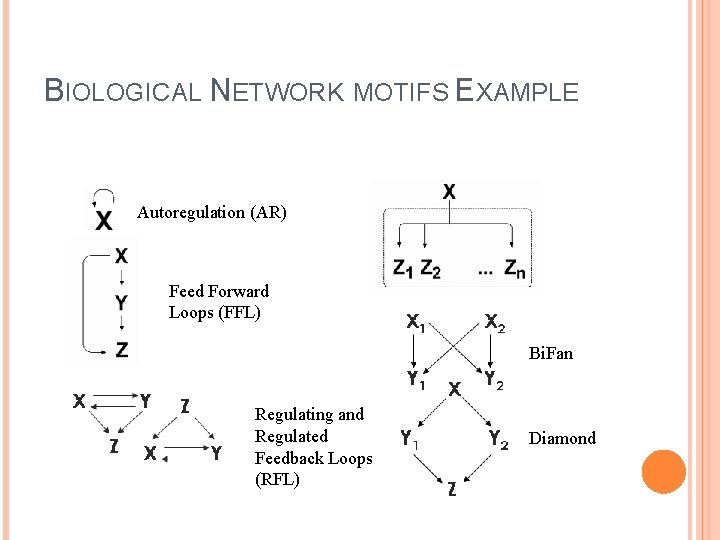
BIOLOGICAL NETWORK MOTIFS EXAMPLE Autoregulation (AR) Feed Forward Loops (FFL) Bi. Fan Regulating and Regulated Feedback Loops (RFL) Diamond

OUTLINE Preliminary Previous Work Method Experimental Conclusion Result

PREVIOUS WORK exhaustive search algorithm � runtime increase dramatically for subgraphs with size ≥ 4. � Impractical to find high-order motifs because of its time complexity. random sampling algorithm � method improves the running time � only estimate the frequency of subgraphs cannot provide an exact solution

OUTLINE Preliminary Previous Work Method Experimental Conclusion Result

METHOD Goal: Find motif from a given graph G(V, E) One Master Processor � Sort all nodes by degree � Partition nodes to Slave processors Slave Processors � Finding Neighborhoods from a Network � Finding Subgraphs within Neighborhood � Gather subgraph set to Master Processor

FINDING NEIGHBORHOODS FROM A NETWORK

FINDING NEIGHBORHOODS FROM A NETWORK

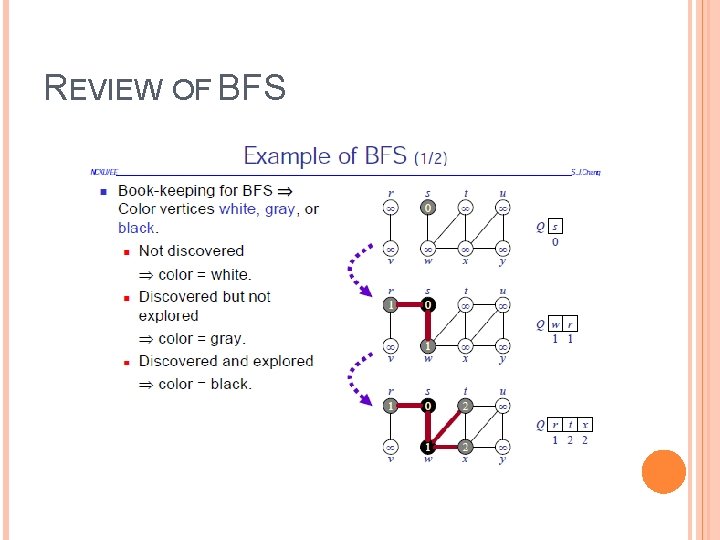
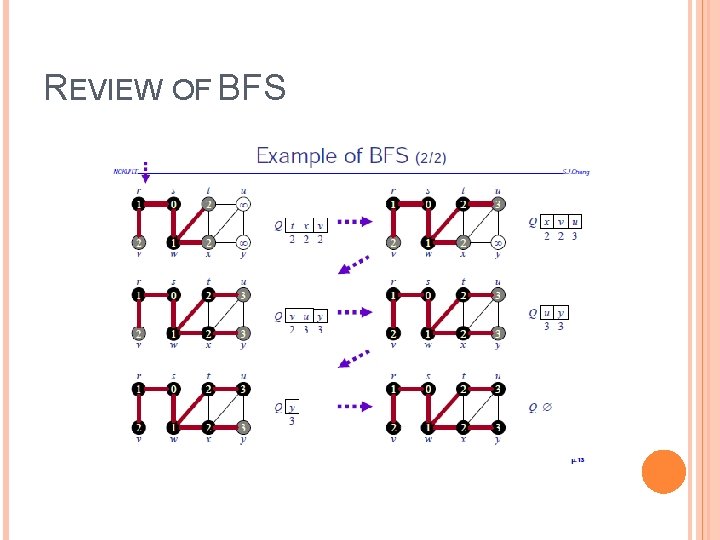
REVIEW OF BFS

REVIEW OF BFS

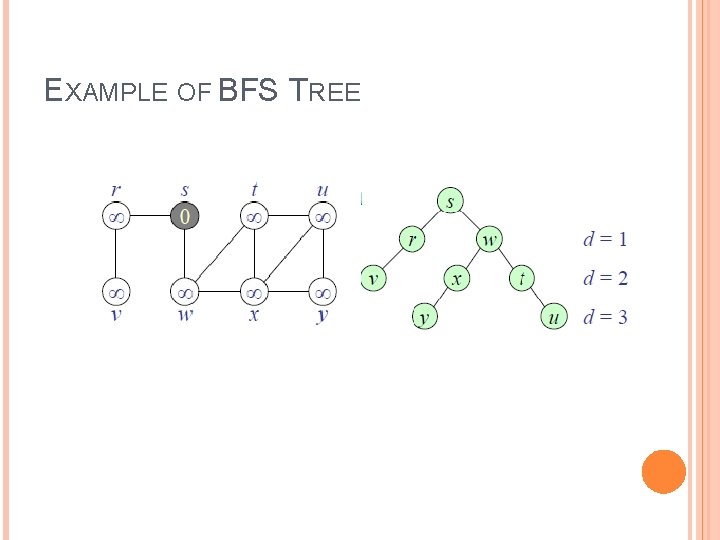
EXAMPLE OF BFS TREE

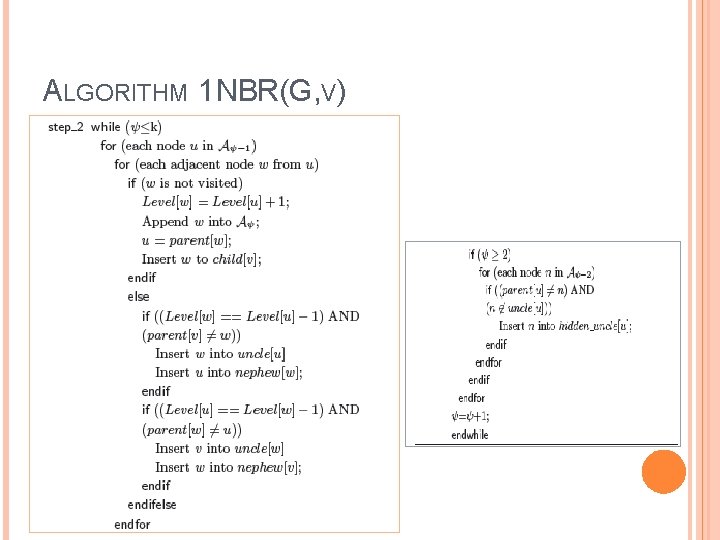
ALGORITHM 1 NBR(G, V)

ALGORITHM 1 NBR(G, V)

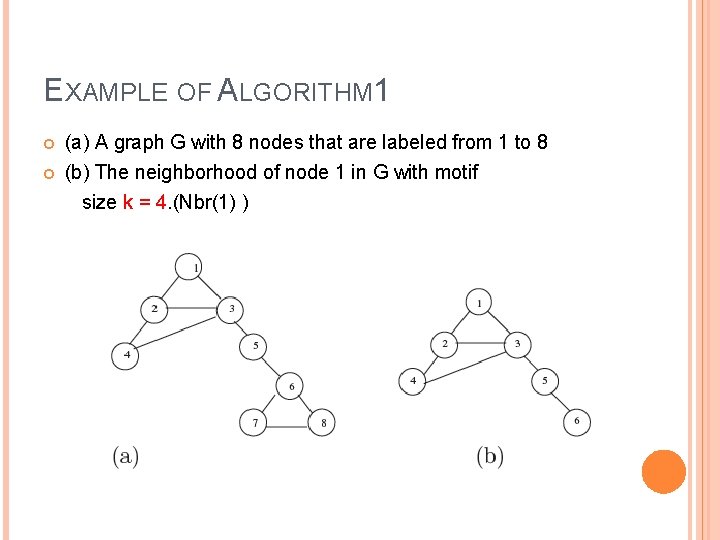
EXAMPLE OF ALGORITHM 1 (a) A graph G with 8 nodes that are labeled from 1 to 8 (b) The neighborhood of node 1 in G with motif size k = 4. (Nbr(1) )


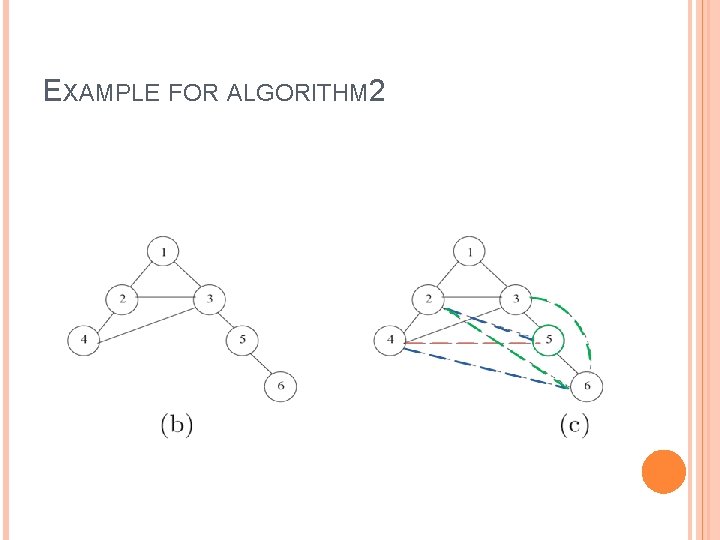
EXAMPLE FOR ALGORITHM 2


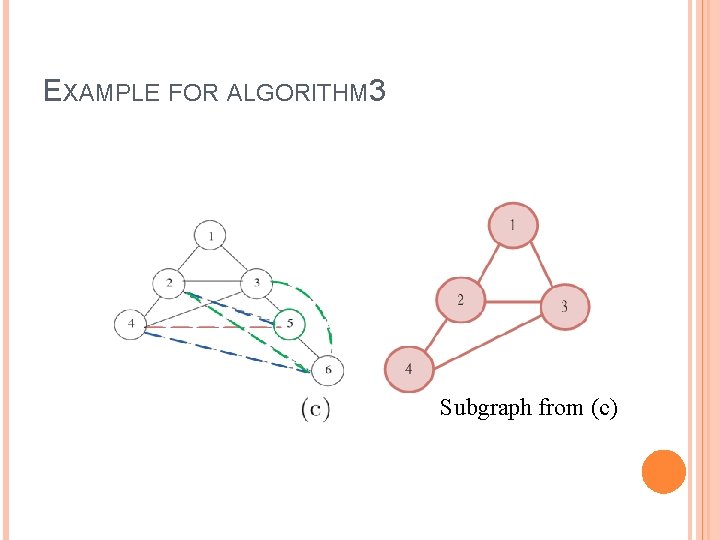
EXAMPLE FOR ALGORITHM 3 Subgraph from (c)


OUTLINE Preliminary Previous Work Method Experimental Conclusion Result

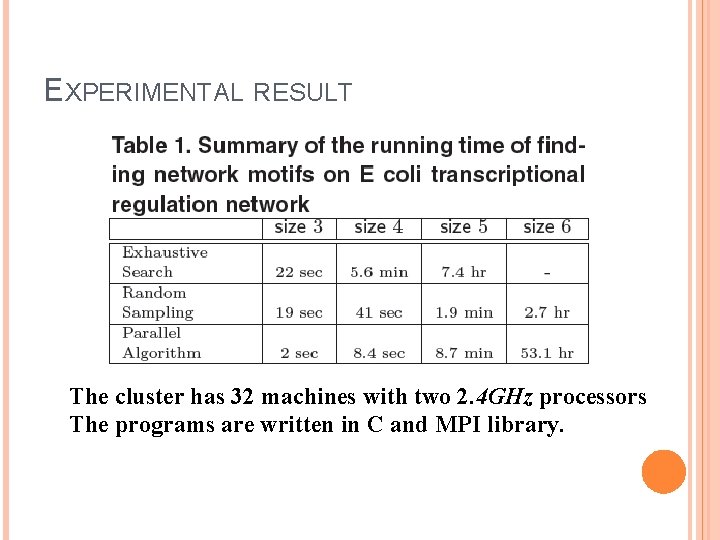
EXPERIMENTAL RESULT The cluster has 32 machines with two 2. 4 GHz processors The programs are written in C and MPI library.

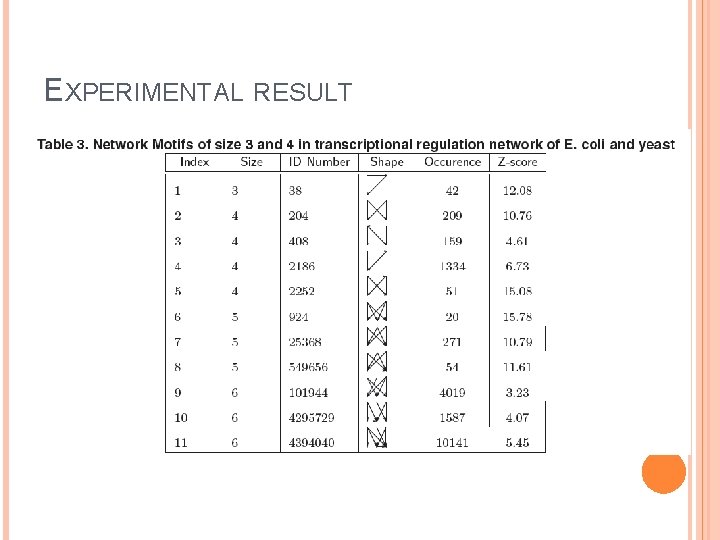
EXPERIMENTAL RESULT Real data set of interactions between transcription factors and operons in an E. coli network from the Regulon. DB database Each protein complex of a transcription factor or a gene is represented by a node.

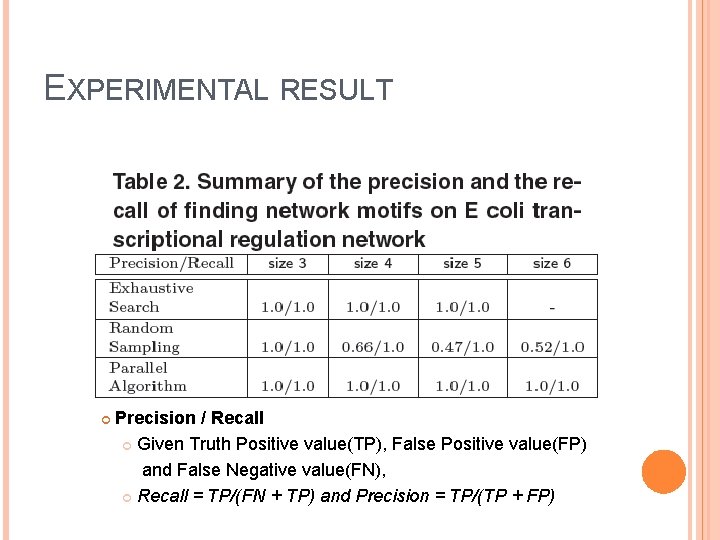
EXPERIMENTAL RESULT Precision / Recall Given Truth Positive value(TP), False Positive value(FP) and False Negative value(FN), Recall = TP/(FN + TP) and Precision = TP/(TP + FP)

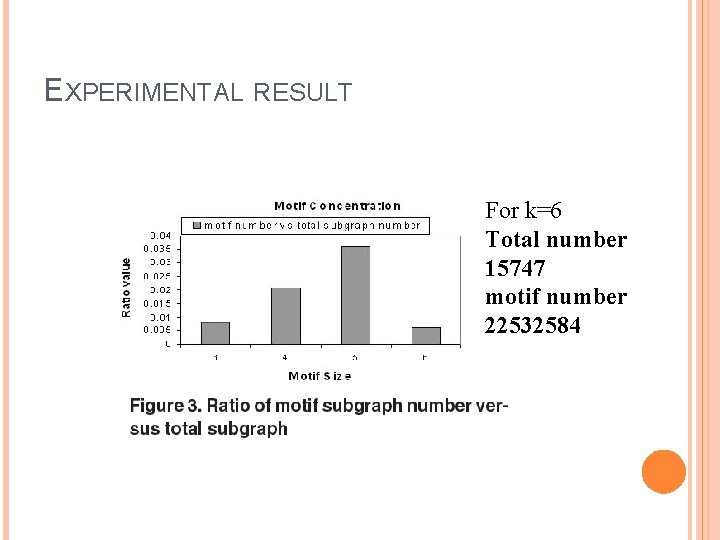
EXPERIMENTAL RESULT For k=6 Total number 15747 motif number 22532584

EXPERIMENTAL RESULT

OUTLINE Preliminary Previous Work Method Experimental Conclusion Result

CONCLUSION This parallel algorithm can accurately find all high-order network motifs in a fast running time. High-order motifs provide important information on biological system design.