A Grid Problem Solving Environment for Bioinformatics the

A Grid Problem Solving Environment for Bioinformatics: the LIBI experience Italo Epicoco, Maria Mirto Research Unit: SPACI Consortium/University of Salento - Lecce Prof. Giovanni Aloisio Martina Franca, Sept. 17 2008, EMBnet Conference - Grid Computing Tutorial

Outline q Grid Problem Solving Environment q The LIBI Project § Meta Scheduler § Grid Portal § Workflow Management System q Applications § Mr. Bayes § Pat. Search § PSI-BLAST q Conclusions and Future Work
What is a Grid Problem Solving Environment? q “A PSE is a computer system that provides all the computational facility needed to solve a target class of problems. . PSEs use the language of the target class of problems. ” (Gallopoulos et al. 1997) q Main components of a PSE are defined in the following equation: § PSE = Natural Language + Solvers + Intelligence + Software Bus q A Grid PSE allows sharing of a heterogeneous set of resources including people, computers, data, and information within an integrated environment (“Virtual Organization”), to solve a target class of problems. Software components of a Grid PSE are exposed as services, grouped in a Grid middleware, and accessible by the users through an easy-to-use interface, named Grid Portal.
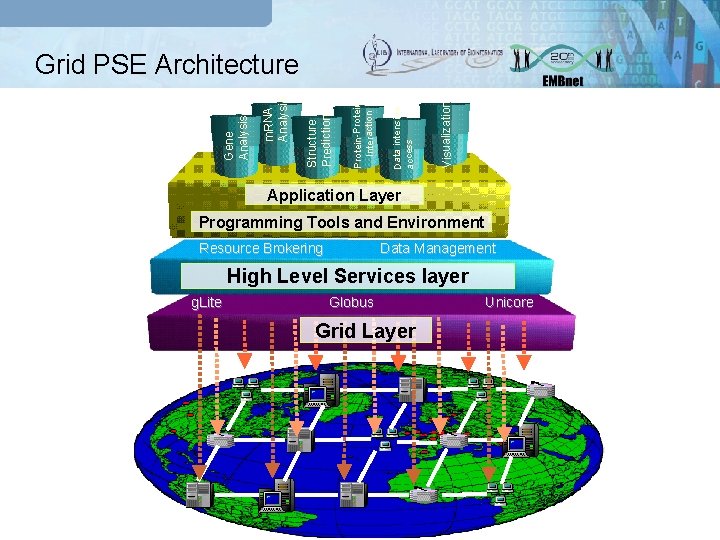
Visualization Data intensive access Protein-Protein Interaction Structure Prediction m. RNA Analysis Gene Analysis Grid PSE Architecture Application Layer Programming Tools and Environment Resource Brokering Data Management High Level Services layer g. Lite Globus Grid Layer Unicore

Grid PSE components Grid Middleware services are offered by several toolkit and they represent the “low level” services for many grid applications. Several examples of Grid middleware are: q Globus; q g. Lite; q Unicore Workflow Service specifies complex (distributed) applications, integrating and composing individual simple services: q Workflow definition through a formal description; q Workflow execution (scheduler) responsible of the enactments, by controlling and coordinating execution of applications. Grid Portal primary requirement is to provide an environment where the user can access Grid resources and services, execute and monitor Grid applications, and collaborate with other users.

The LIBI Grid PSE q Several goals: § Building of a technological framework that, guaranting the interoperability between different grid middleware (g. Lite, Unicore, Globus), provides an environment for composing, executing and monitoring complex experiments in Bioinformatics; § Optimization and porting in the Grid of Bioinformatics applications.
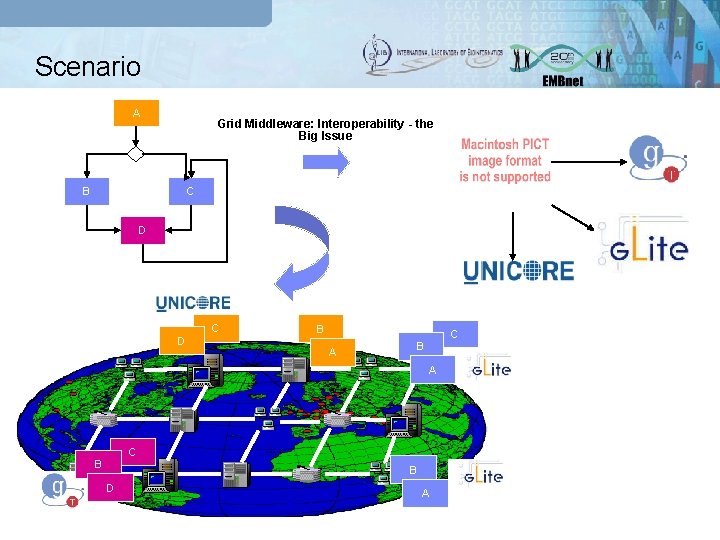
Scenario A Grid Middleware: Interoperability - the Big Issue B C D D C B A A C B B D A

Meta Scheduler q We have developed a Web Service component that supports the submission and the monitoring of workflows, batch, MPI and parameter sweep jobs distributed on g. Lite, Globus and Unicore based grids; q Several libraries, plugged in the meta scheduler, provide core functions for job submission/monitoring and data transfer by using the Grid. FTP protocol; q Used JSDL specification; q Automatic converters from JSDL into specific grid languages for the submission; q Support for the applications wrapped as Web Services (work in progress).
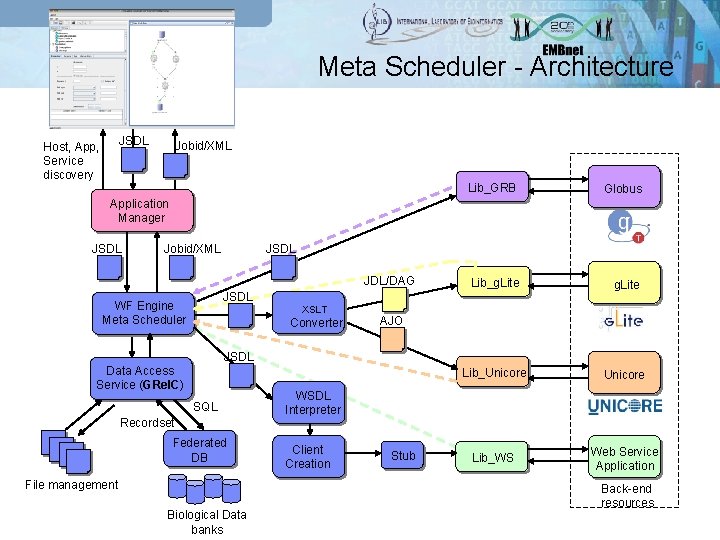
Meta Scheduler - Architecture JSDL Host, App, Service discovery Jobid/XML Lib_GRB Globus Lib_g. Lite Lib_Unicore Lib_WS Web Service Application Manager JSDL Jobid/XML JSDL JDL/DAG JSDL WF Engine Meta Scheduler XSLT Converter AJO JSDL Data Access Service (GRel. C) SQL Recordset Federated DB File management Biological Data banks WSDL Interpreter Client Creation Stub Back-end resources

Job submission: different approaches q Meta scheduler Job Submission: § Stage in of needed input files § Stage in of the executable § Job execution § Stage out of produced output files q Globus § Using RSL we can define once the file to be staged in, execute and staged out. The file transfer is made by GRAM q g. Lite § The job is specified through a JDL description this can include references to remote files to be staged in or staged out from SE § The files must be present in the SE before job starting. The file transfer must be made explicitly by the client through a gridftp session q UNICORE § The job is specified through an AJO object § A single AJO does not contain references to remote files to be transferred § The stage in of stage out phase must be made explicitly by the client submitting a job for file transfer

Job support q Meta scheduler job types: § Single batch: synchronous and asynchronous § Parameter sweep job § Workflow job q Globus § Does not support parameter sweep nor workflow job q g. Lite § Supports job array, workflow job (only DAG), batch job q UNICORE § Does not support parameter sweep jobs

Security q Meta scheduler Security § Mutual authentication § Single sign on § Support for credential delegation q Globus § Based on GSI, it supports mutual authentication, single sign on, delegation q g. Lite § Based on GSI, it supports the VO basis authorization extension q UNICORE § Supports GSI, but the credential delegation not yet

Job tracking q Meta Scheduler Job Tracking: § Tracking § User notification q Globus § GRAM takes care about the status of the job. § It does not support notification. § The returned job status is not reliable q g. Lite § LB takes care about the status of the job § It does not support notification. § The returned job status is not reliable q UNICORE § NJS takes care about the status of the job § It does not support notification.
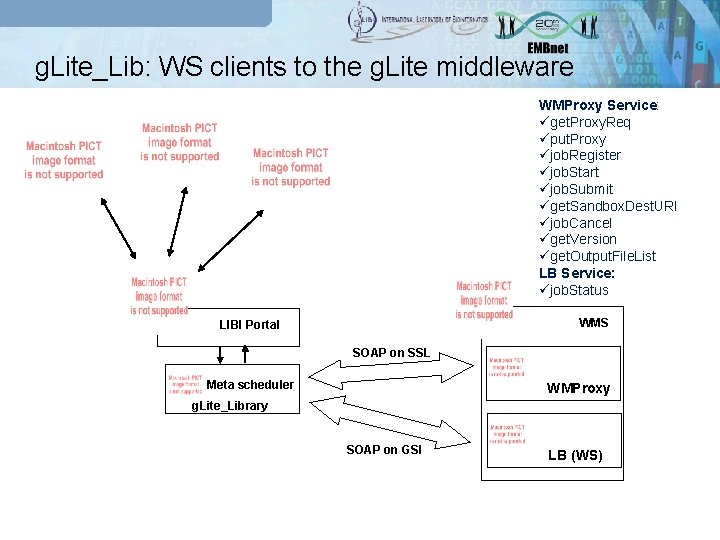
g. Lite_Lib: WS clients to the g. Lite middleware WMProxy Service: üget. Proxy. Req üput. Proxy üjob. Register üjob. Start üjob. Submit üget. Sandbox. Dest. URI üjob. Cancel üget. Version üget. Output. File. List LB Service: üjob. Status WMS LIBI Portal SOAP on SSL Meta scheduler WMProxy g. Lite_Library SOAP on GSI LB (WS)
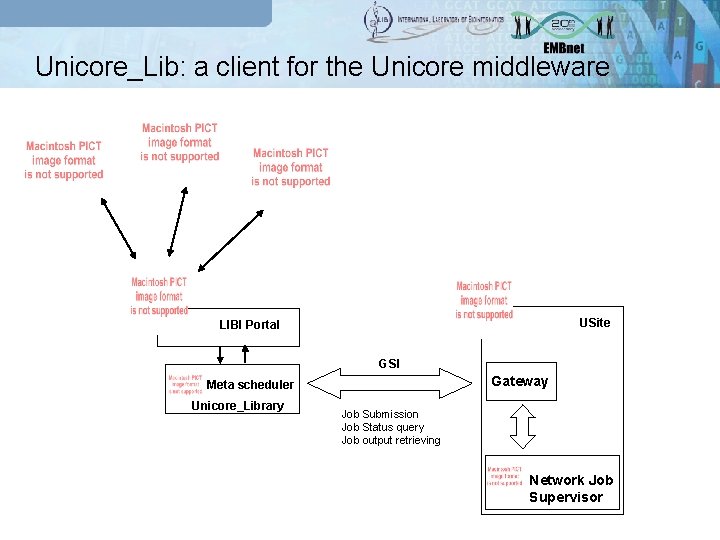
Unicore_Lib: a client for the Unicore middleware USite LIBI Portal GSI Gateway Meta scheduler Unicore_Library Job Submission Job Status query Job output retrieving Network Job Supervisor

LIBI Grid Portal - Features q Based on GRB technology, it allows the management of the following information: § Profile § Credential § VO Management § Resource Management § Database Management § Software Management § View Configuration
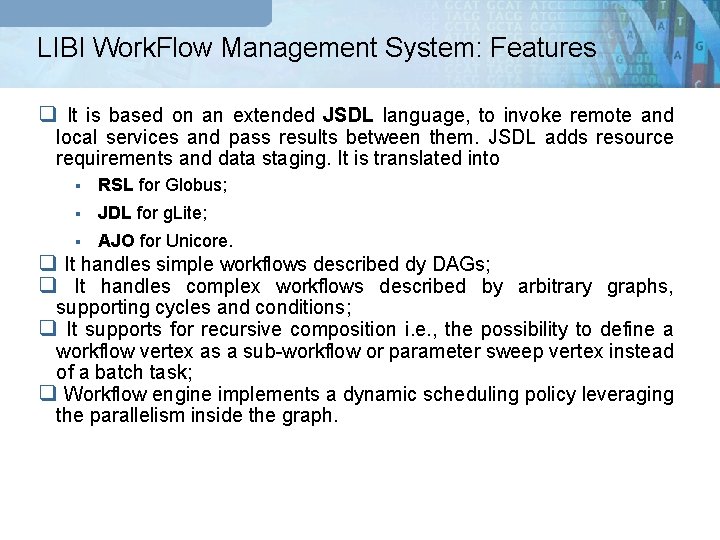
LIBI Work. Flow Management System: Features q It is based on an extended JSDL language, to invoke remote and local services and pass results between them. JSDL adds resource requirements and data staging. It is translated into § RSL for Globus; § JDL for g. Lite; § AJO for Unicore. q It handles simple workflows described dy DAGs; q It handles complex workflows described by arbitrary graphs, supporting cycles and conditions; q It supports for recursive composition i. e. , the possibility to define a workflow vertex as a sub-workflow or parameter sweep vertex instead of a batch task; q Workflow engine implements a dynamic scheduling policy leveraging the parallelism inside the graph.

Taverna and Bio-STEER q Taverna: is a workflow orchestration tool for web services and it is part of the my. Grid project. § It can handle WSDL based web service invocation, and query a standard UDDI registry, taking into account user’s preferences to obtain actual service instances to invoke. § It supports the XML-based workflow definition language named SCUFL (Simple Conceptual Unified Flow Language), to invoke remote and local services and pass results between them. § The disadvantage of Taverna is that the user can not define complex operations such as loops or conditions and it has issues for managing huge amount of data q Bio-STEER: provides an environment in which the user can compose custom workflows. § At each step, Bio-STEER utilizes the application semantics (particularly the input and output types) in order to assist the user in narrowing down the list of potential services that can go into the workflow. § It uses an ontology and supports cycles and conditions; long running processes and parallel processing are also handled. § Bio-STEER allows the composition of web services only and does not allow the execution on different grid middleware.
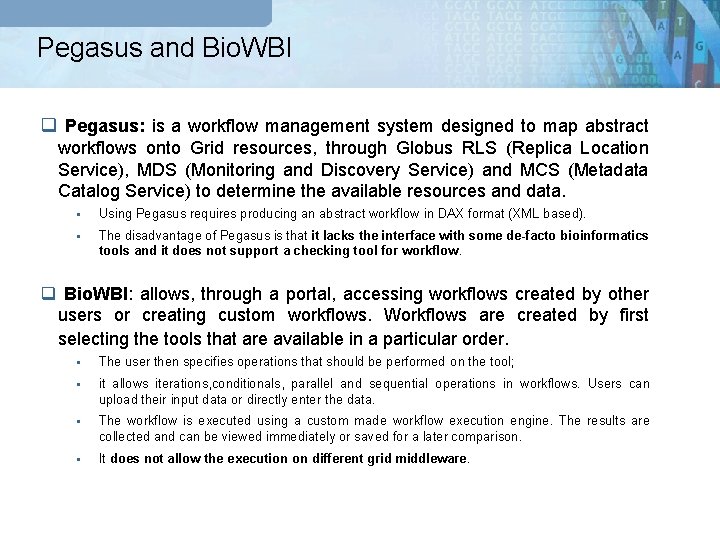
Pegasus and Bio. WBI q Pegasus: is a workflow management system designed to map abstract workflows onto Grid resources, through Globus RLS (Replica Location Service), MDS (Monitoring and Discovery Service) and MCS (Metadata Catalog Service) to determine the available resources and data. § Using Pegasus requires producing an abstract workflow in DAX format (XML based). § The disadvantage of Pegasus is that it lacks the interface with some de-facto bioinformatics tools and it does not support a checking tool for workflow. q Bio. WBI: allows, through a portal, accessing workflows created by other users or creating custom workflows. Workflows are created by first selecting the tools that are available in a particular order. § The user then specifies operations that should be performed on the tool; § it allows iterations, conditionals, parallel and sequential operations in workflows. Users can upload their input data or directly enter the data. § The workflow is executed using a custom made workflow execution engine. The results are collected and can be viewed immediately or saved for a later comparison. § It does not allow the execution on different grid middleware.
Pegasys and Condor DAG - Man q Pegasys: is a workflow management system for bioinformatics. § The system includes numerous tools for pairwise and multiple sequence alignment, ab initio gene prediction, RNA gene detection, etc. § It allows building direct acyclic graphs. § It does not exploit a Grid framework and must be installed on a cluster machine. q Condor DAGMan: supports execution of simple workflows described by DAGs through its DAGMan component, which can be thought as a task sequencing engine. § Ready jobs are submitted from DAGMan to the Condor scheduler taking into account the data dependencies in the workflow. § The scheduler selects computing resources matching the jobs in terms of requirements interacting with a negotiator module and submits the jobs. § A job status is then passed back to DAGMan which monitors the status of all of the workflow jobs running in order to update dynamically the ready jobs queue. § The user can not define complex operations such as loops or conditions.

Grid. Way is a meta scheduler able to submit and monitor jobs on both Globus and g. Lite grids. The main differences, with regard to our work are the following ones: • • • Grid. Way does not submit jobs on UNICORE grids; Grid. Way requires all of the user accounts it acts on behalf of to be available on the same machine where it is installed. The reason is that Grid-Way switches at runtime its unix execution id with the id of the user it needs to submit on behalf of. Our workflow management system does not require this, owing to the fact that it uses an external My. Proxy server to obtain X 509 v 3 digital certificates as credentials related to the users; Grid. Way supports the DRMA (Distributed Resource Management Application) API, but does not provide support for management of jobs described by complex workflow including cycles etc; a basic support for DAG based jobs however, is available.

LIBI WFMS: GUI q An XML-based language for specifying the job flow for biological applications (abstract workflow). It describes graphical elements; q Discovery of available tools into the LIBI Grid PSE; q For each application it is possible to specify arguments, processors number (for MPI jobs), stdin, stdout, stderr. q Submitted workflow are saved with a logical name; q Visualization, updating and saving of the graph on the Web Server; q Zoom-in, zoom-out of the graph and save it in several formats (jpeg, png, gif).

Workflow Applet

Grid PSE: Applications Services (1/2) q Mr. Bayes for Bayesian estimation of phylogeny – test made on Globus based resources – MPI jobs q Pat. Search for retrieving patterns into a sequence – test made on g. Lite resources – Batch and parameter sweep jobs q PSI-BLAST for multiple sequence alignment – test made on Globus based resources – Workflow job that involves a job for extracting sequences by input files in different formats and another job for running the same application several times with different inputs

Grid PSE: Applications Services (2/2) q Gromacs for proteins dynamics simulation – test made on Globus and Unicore based resources – MPI jobs q Antihunter for the identification of expressed sequence tag (EST) antisense transcripts from BLAST output – test made on Unicore based resources – Batch jobs – A case study in the Falciano’s talk
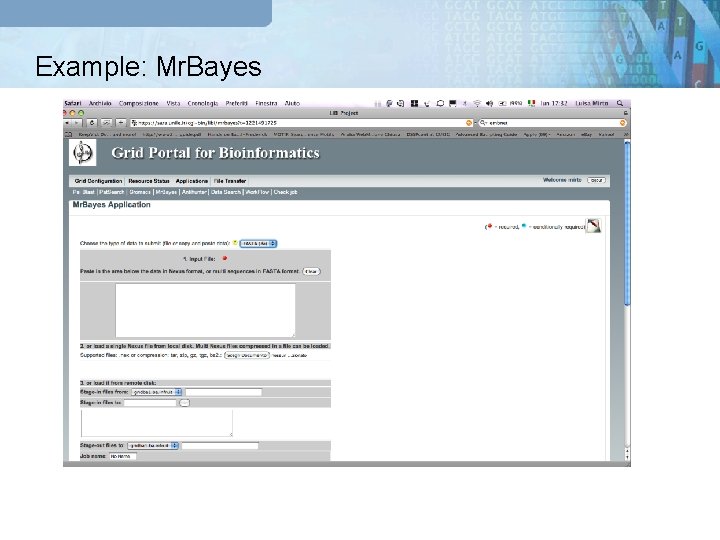
Example: Mr. Bayes GRB Portal Meta Scheduler Globus GT 4 g. Lite UNICORE
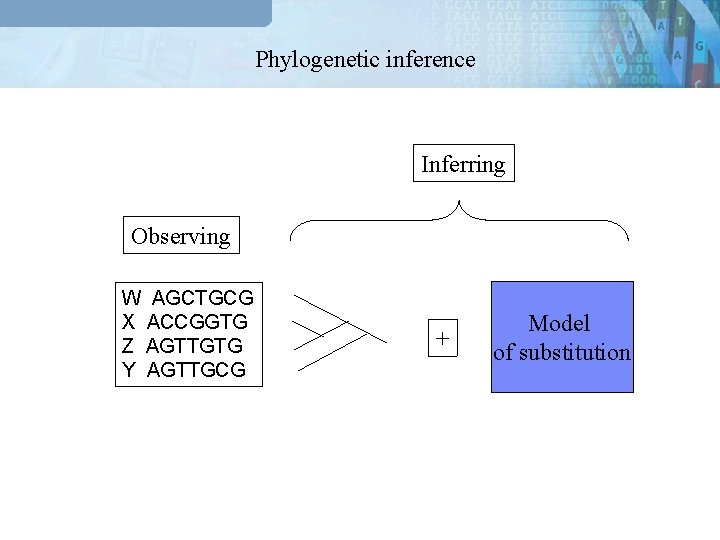
Phylogenetic inference Inferring Observing W AGCTGCG X ACCGGTG Z AGTTGTG Y AGTTGCG + Model of substitution

Very complex analytical solution = Bayesian formula cannot be used because is too long. The integration of the likelihood also cannot be estimate directly because is too difficult. Then. . ? Markovian integration
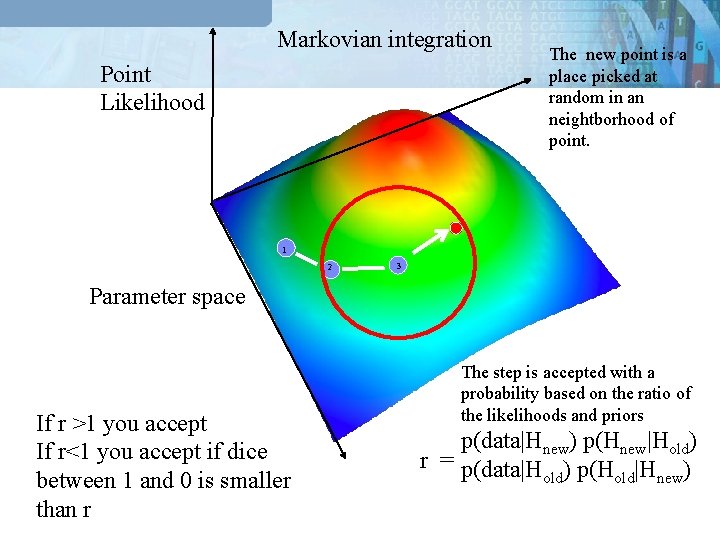
Markovian integration Point Likelihood The new point is a place picked at random in an neightborhood of point. 1 2 3 Parameter space If r >1 you accept If r<1 you accept if dice between 1 and 0 is smaller than r The step is accepted with a probability based on the ratio of the likelihoods and priors p(data|Hnew) p(Hnew|Hold) r = p(data|H ) p(H |H ) old new
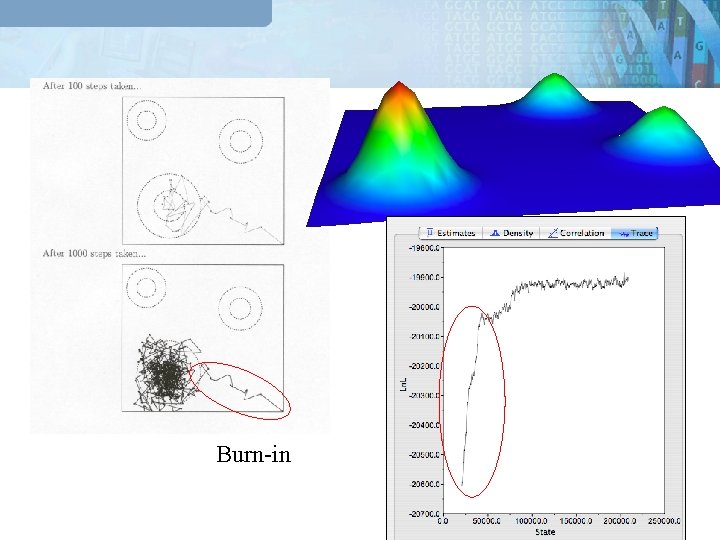
Burn-in
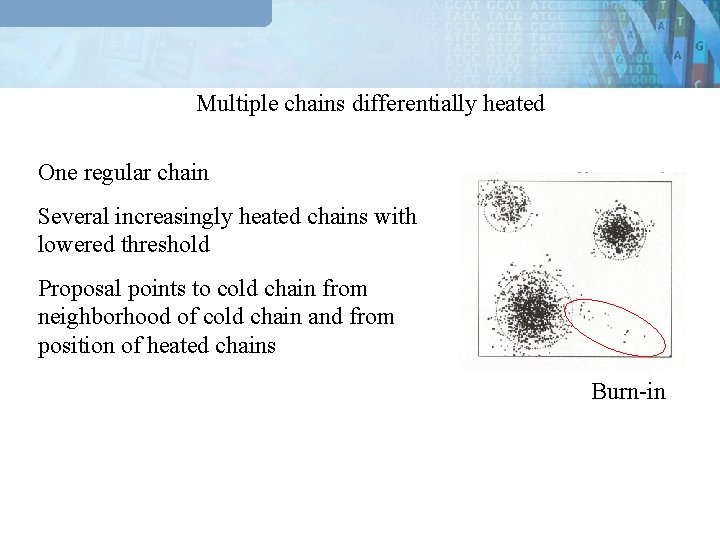
Multiple chains differentially heated One regular chain Several increasingly heated chains with lowered threshold Proposal points to cold chain from neighborhood of cold chain and from position of heated chains Burn-in

And how I get the topology estimation What is the probability given the data that taxa X is sister taxa of taxon W What percentage of steps have taxa X sister taxa of taxa W
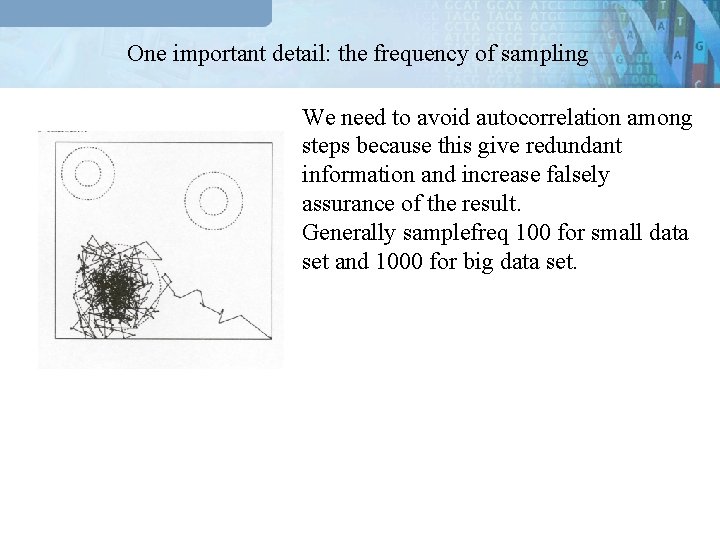
One important detail: the frequency of sampling We need to avoid autocorrelation among steps because this give redundant information and increase falsely assurance of the result. Generally samplefreq 100 for small data set and 1000 for big data set.

How to access LIBI Grid PSE q Contact URL: § https: //sara. unile. it/cgi-bin/libi/enter q Available accounts: § Username: test. X § Passwd: test. X where X=1, 2, . . , 10

Mr. Bayes Case study q Two options as input: § File nexus § File FASTA 1. Please download the file at the following url: http: //sara. unile. it/libi/demo_libi/mrbayes/Cervus. COMP. cfas. afa. tgz 2. Uncompress the file and upload it from the portal https: //sara. unile. it/cgi-bin/libi/enter Help on line: http: //sara. unile. it/libi/tutorial_mrbayes. html
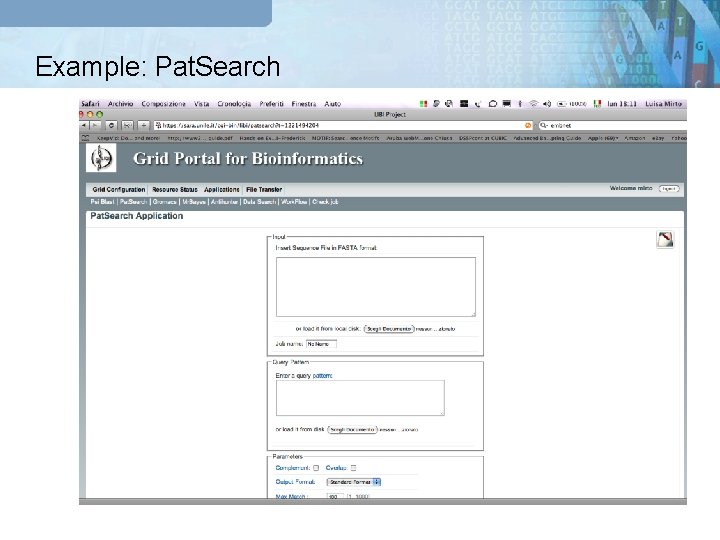
Example: Pat. Search GRB Portal Meta Scheduler Globus GT 4 g. Lite UNICORE

Pat. Search Case study q Two file as input: § FASTA file contains the sequences: fasta § CMD file: input_cmd 1. Please download the file at the following url: http: //sara. unile. it/libi/demo_libi/patsearch_input. tgz 2. Uncompress the file and upload it from the portal https: //sara. unile. it/cgi-bin/libi/enter
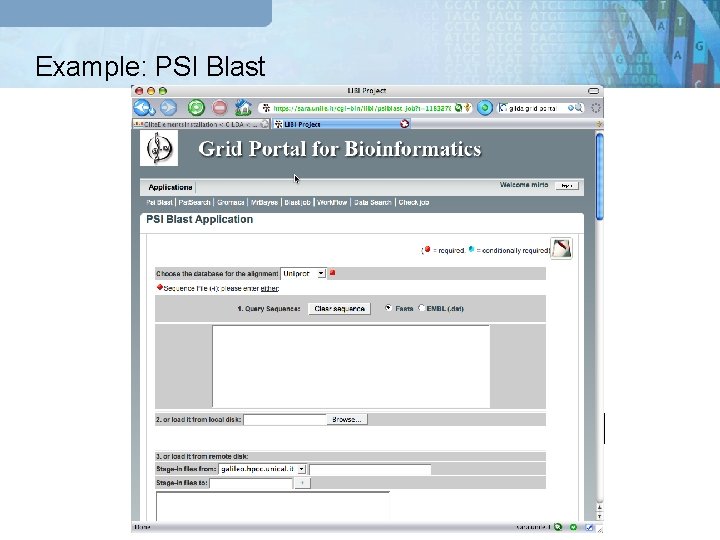
Example: PSI Blast GRB Portal Meta Scheduler Globus GT 4 g. Lite UNICORE
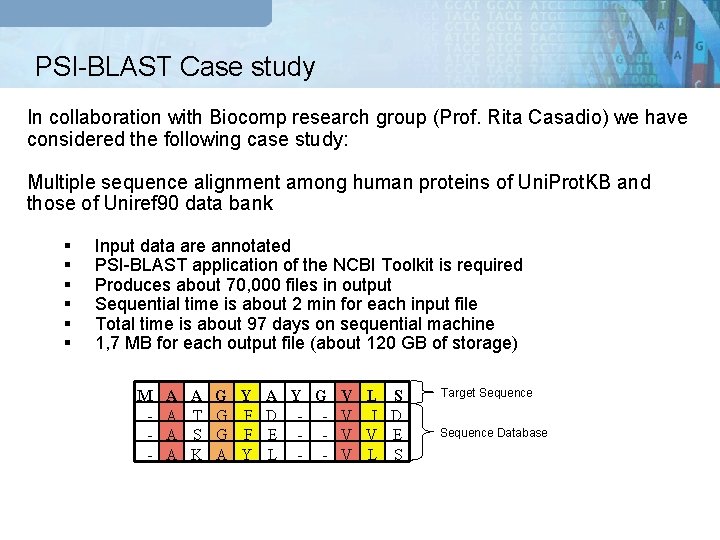
PSI-BLAST Case study In collaboration with Biocomp research group (Prof. Rita Casadio) we have considered the following case study: Multiple sequence alignment among human proteins of Uni. Prot. KB and those of Uniref 90 data bank § § § Input data are annotated PSI-BLAST application of the NCBI Toolkit is required Produces about 70, 000 files in output Sequential time is about 2 min for each input file Total time is about 97 days on sequential machine 1, 7 MB for each output file (about 120 GB of storage) M - A A G Y A T G F A S G F A K A Y G V L S D - - V I D E - - V V E L - - V L S Target Sequence Database
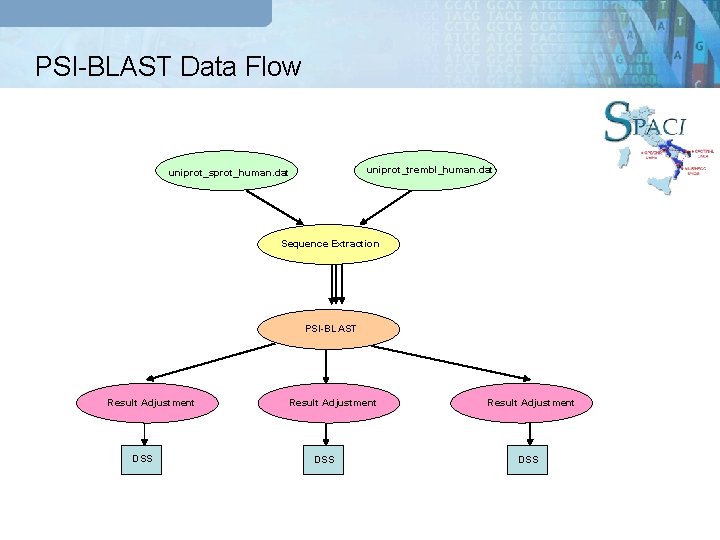
PSI-BLAST Data Flow uniprot_trembl_human. dat uniprot_sprot_human. dat Sequence Extraction PSI-BLAST Result Adjustment DSS

PSI-BLAST Case study q Two kind of input: § FASTA file or § DAT file (annotated file) 1. Please download the file at the following url: http: //sara. unile. it/libi/demo_libi/psiblast/uniprot_sprot_human. dat_10. tgz 2. Uncompress the file and upload it from the portal https: //sara. unile. it/cgi-bin/libi/enter Help on line: http: //sara. unile. it/libi/tutorial_psiblast. html

Conclusions and Future Works q Integration of the Job Submission Tool (JST) Donvito’s talk - in the Meta scheduler; q Development of a plugin for the support to the CREAM and ICE service of g. Lite; q Development of the jobs monitoring into the GUI; q Test and deployment of the WFMS.

SPACI - UNILE Team q Prof. Giovanni Aloisio (Supervisor) § giovanni. aloisio@unile. it q Maria Mirto § maria. mirto@unile. it q Italo Epicoco § italo. epicoco@unile. it q Massimo Cafaro § massimo. cafaro@unile. it q Alessandro D’Anca § alessandro. danca@unile. it q Alessandro Negro § alessandro. negro@unile. it q Daniele Tartarini § daniele. tartarini@unile. it q Marco Passante § marco. passante@libero. it
- Slides: 44