A data retrieval workflow using NCBI EUtils Python

A data retrieval workflow using NCBI E-Utils + Python Part II: Jinja 2 / Flask John Pinney Tech talk Tue 19 th Nov

My tasks ✓ 1. Produce a list of human genes that are associated with at least one resolved structure in PDB AND at least one genetic disorder in OMIM 2. Make an online table to display them
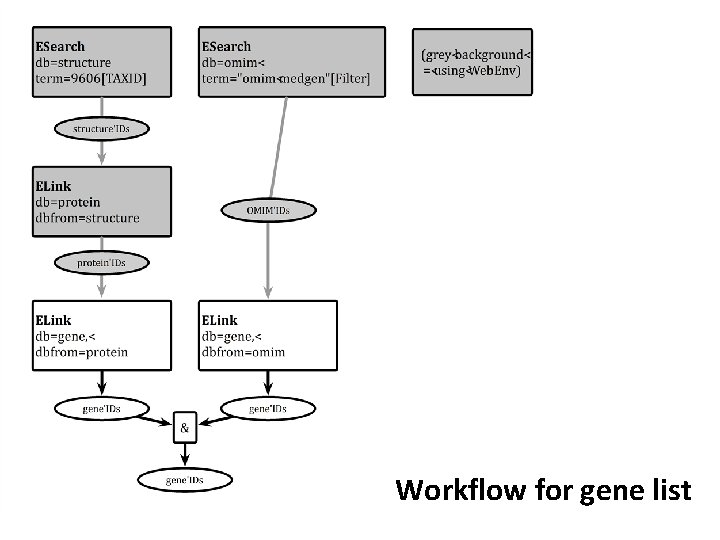
Workflow for gene list

Python modules used in part 1 Py. Cogent Simple request handling for the main EUtils. pycogent. org urllib 2 General HTTP request handler. docs. python. org/2/library/urllib 2. html Beautiful. Soup Amazingly easy to use object model for XML/HTML. www. crummy. com/software/Beautiful. Soup/bs 4/doc/

Some REST services need API keys The OMIM server requires a license agreement but is free for academic use. They provide a personal API key which must be submitted with each HTTP request. OMIM_APIKEY = 'E 835870 B 16 FBAF 479 E 826 FA 5168 CB 2615 EDA 0 F 11' result = urllib 2. urlopen( "http: //api. europe. omim. org/api/entry? mim. Number=" + omimid + "&api. Key=" + OMIM_APIKEY ). read()

Throttling queries Most bioinformatics web servers have limits on the number of queries that can be sent from the same IP address (per day / per second etc. ) They will ban you from accessing the site if you attempt too many requests. This can have serious consequences (e. g. the whole institution being blocked from NCBI).
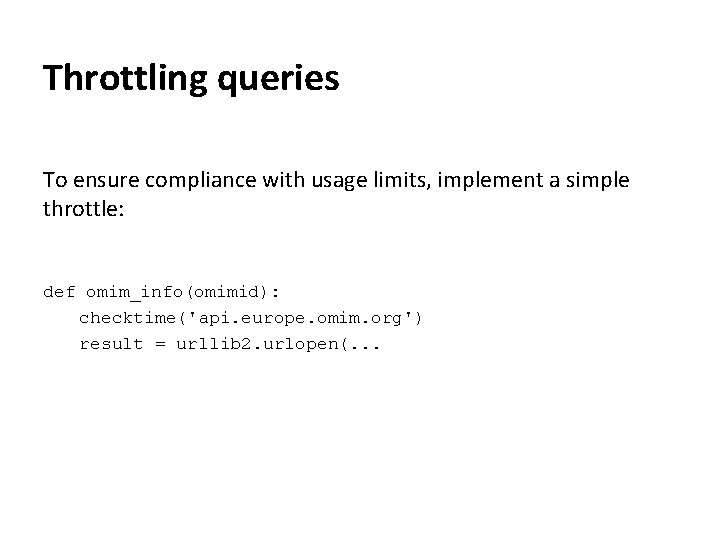
Throttling queries To ensure compliance with usage limits, implement a simple throttle: def omim_info(omimid): checktime('api. europe. omim. org') result = urllib 2. urlopen(. . .

Throttling queries import time last. Request. Time = {} throttle. Delay = {'eutils. ncbi. nlm. nih. gov': 0. 25, 'api. europe. omim. org': 0. 5} def checktime(host): if((host in last. Request. Time) and (time() - last. Request. Time[host] < throttle. Delay[host])): time. sleep(throttle. Delay[host] - (time() - last. Request. Time[host])) last. Request. Time[host] = time()
HTML templating I need to produce an HTML table containing basic information about the genes I have collected. The Jinja 2 templating engine is an easy way to generate these kinds of documents. I will use web services at NCBI and OMIM to assemble the information I need.

Jinja 2 Using Jinja 2 as an HTML templating engine, we need to split the work between 2 files: a normal python script (in which I call the web services). an HTML template with embedded python commands. Not all python functions are available within the template, so it makes sense to do as much work as possible within the script before passing the data over.

Jinja 2 (script) from jinja 2 import Template template = Template(file("gene_row_template. html"). read()) fout = open("gene_list. html", 'w'). . . (variables passed to template as for g in sorted_genes: kwargs) fout. write( template. render( g=g, gene=gene_info(g), omim=[omim_info(x) for x in omim_links(g)], struc=[struc_info(x) for x in struc_links(g)] ) )
![Jinja 2 (template) <tr> <td><a href='http: //www. ncbi. nlm. nih. gov/gene/? term={{g}}[uid]'> {{gene. find('Gene-ref_locus'). Jinja 2 (template) <tr> <td><a href='http: //www. ncbi. nlm. nih. gov/gene/? term={{g}}[uid]'> {{gene. find('Gene-ref_locus').](http://slidetodoc.com/presentation_image_h2/f85ec65db7f8059183fb6b12c0e26aa8/image-12.jpg)
Jinja 2 (template) <tr> <td><a href='http: //www. ncbi. nlm. nih. gov/gene/? term={{g}}[uid]'> {{gene. find('Gene-ref_locus'). text}} </a></td> <td>{{gene. find('Gene-ref_desc'). text}}</td> <td>{% for m in omim %} <a href='http: //omim. org/entry/{{m. mim. Number. text}}'> {{m. preferred. Title. text}} </a> {% endfor %}</td> <td>{% for s in struc -%} <a href='http: //www. rcsb. org/pdb/explore. do? structure. Id={{s. find(' Item', attrs={'Name': 'Pdb. Acc'}). text}}'> {{s. find('Item', attrs={'Name': 'Pdb. Acc'}). text}} </a> {%- endfor %}</td> </tr>
![Jinja 2 (template) <tr> <td><a href='http: //www. ncbi. nlm. nih. gov/gene/? term={{g}}[uid]'> {{gene. find('Gene-ref_locus'). Jinja 2 (template) <tr> <td><a href='http: //www. ncbi. nlm. nih. gov/gene/? term={{g}}[uid]'> {{gene. find('Gene-ref_locus').](http://slidetodoc.com/presentation_image_h2/f85ec65db7f8059183fb6b12c0e26aa8/image-13.jpg)
Jinja 2 (template) <tr> <td><a href='http: //www. ncbi. nlm. nih. gov/gene/? term={{g}}[uid]'> {{gene. find('Gene-ref_locus'). text}} {{ }} = print statement </a></td> <td>{{gene. find('Gene-ref_desc'). text}}</td> <td>{% for m in omim %} <a href='http: //omim. org/entry/{{m. mim. Number. text}}'> {{m. preferred. Title. text}} </a> {% %} = other command {% endfor %}</td> <td>{% for s in struc -%} <a href='http: //www. rcsb. org/pdb/explore. do? structure. Id={{s. find(' Item', attrs={'Name': 'Pdb. Acc'}). text}}'> {{s. find('Item', attrs={'Name': 'Pdb. Acc'}). text}} </a> I can access the methods of an object from {%- endfor %}</td> within the template, so I can make use of all </tr> the nice Beautiful. Soup shortcuts
![Jinja 2 (output) <tr> <td><a href='http: //www. ncbi. nlm. nih. gov/gene/? term=94[uid]'> ACVRL 1 Jinja 2 (output) <tr> <td><a href='http: //www. ncbi. nlm. nih. gov/gene/? term=94[uid]'> ACVRL 1](http://slidetodoc.com/presentation_image_h2/f85ec65db7f8059183fb6b12c0e26aa8/image-14.jpg)
Jinja 2 (output) <tr> <td><a href='http: //www. ncbi. nlm. nih. gov/gene/? term=94[uid]'> ACVRL 1 </a></td> <td>activin A receptor type II-like 1</td> <a href='http: //omim. org/entry/600376'> TELANGIECTASIA, HEREDITARY HEMORRHAGIC, TYPE 2; HHT 2 </a> </td> <td><a href='http: //www. rcsb. org/pdb/explore. do? structure. Id=4 FAO'> 4 FAO </a> <a href='http: //www. rcsb. org/pdb/explore. do? structure. Id=3 MY 0'> 3 MY 0 </a> </td> </tr>

Something more interactive What if I need to produce a report on-the-fly? Flask is a ‘micro’ web development framework for Python, which is useful for putting together a simple webserver. For anything more substantial (e. g. if database queries are needed), consider using Django. Flask uses Jinja 2 as its template engine.

A simple webapp in Flask from flask import Flask, request, render_template, Response app = Flask(__name__) @app. route('/report/') def report_handler(): gene = request. args. get('gene') if( gene == None): return render_template('report_form. html', unfound=None) else: return report_for_gene_name(gene) if __name__ == '__main__': app. run(debug=True)

Summary Some web services may be more fiddly than others to set up, especially if they involve API keys Request limits (requires throttling) Combining web services with an HTML template (either offline or on-the-fly via a webserver) is an easy way to generate userfriendly reports.

Python modules used in part 2 Jinja 2 An elegant and highly versatile templating engine. http: //jinja. pocoo. org/ Flask Python ‘micro’ web development framework. http: //flask. pocoo. org
- Slides: 18