A computer scientists journey with biomedical computing Dynamics

A computer scientist’s journey with bio-medical computing: Dynamics in Data Abstract: Biomedical imaging is yet to harness the full information contents of dynamics in data. I am not referring to longitudinal data over disease progression, but to real-time dynamics during the imaging, especially in molecular imaging. As a computer scientist our group is exposed to with such real-time dynamic data - from dynamic single photon emission computed tomography (SPECT) image reconstruction to dynamic contrast-enhanced ultrasound (CEUS) image analysis. My talk will try to illustrate a few steps in my group’s journey in bio-medical image computing. This is not a deep medical or mathematical talk, but rather a sales pitch touching upon four or five of our “products” and products-under-development. Debasis Mitra teaches computer science at the Florida Institute of Technology in Melbourne, Florida. Mitra holds Ph. D. in mathematical physics and in computer science. His interests are in artificial intelligence, optimization, and biomedical imaging.

A computer scientist’s “journey” with (read: grab-bag of) biomedical computing Debasis Mitra Florida Institute of Technology
Cs. fit. edu/~dmitra 10/19/2021 D. Mitra at IBIIS, Stanford 3
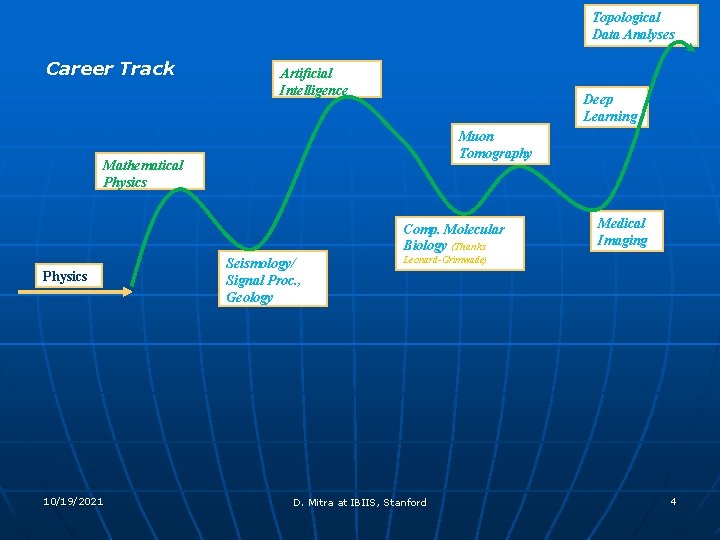
Topological Data Analyses Career Track Artificial Intelligence Deep Learning Muon Tomography Mathematical Physics Comp. Molecular Biology (Thanks Physics 10/19/2021 Seismology/ Signal Proc. , Geology Medical Imaging Leonard-Grimwade) D. Mitra at IBIIS, Stanford 4

Project I Computational Molecular Biology: Protein Tertiary-structure Analyses Stephen Johnsson, Ph. D. 10/19/2021 D. Mitra at IBIIS, Stanford 5
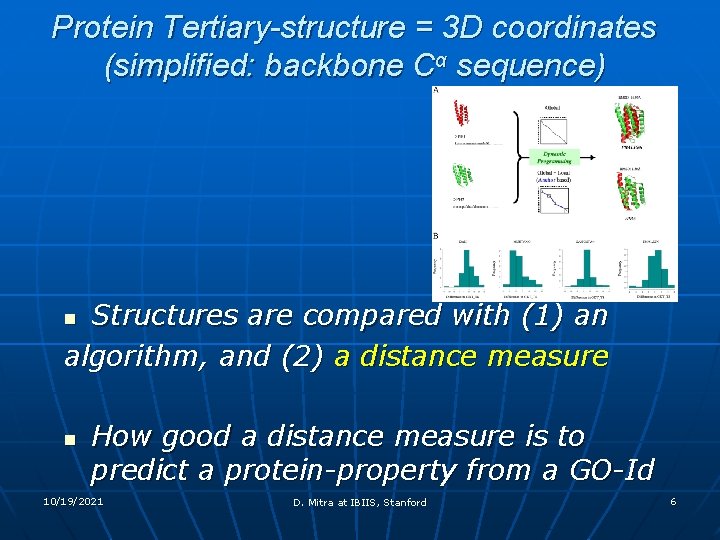
Protein Tertiary-structure = 3 D coordinates (simplified: backbone Cα sequence) Structures are compared with (1) an algorithm, and (2) a distance measure n n How good a distance measure is to predict a protein-property from a GO-Id 10/19/2021 D. Mitra at IBIIS, Stanford 6
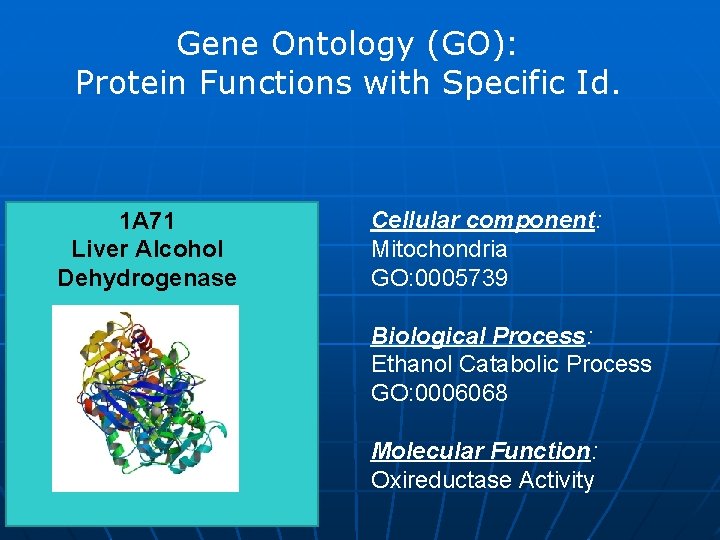
Gene Ontology (GO): Protein Functions with Specific Id. 1 A 71 Liver Alcohol Dehydrogenase Cellular component: Mitochondria GO: 0005739 Biological Process: Ethanol Catabolic Process GO: 0006068 Molecular Function: Oxireductase Activity
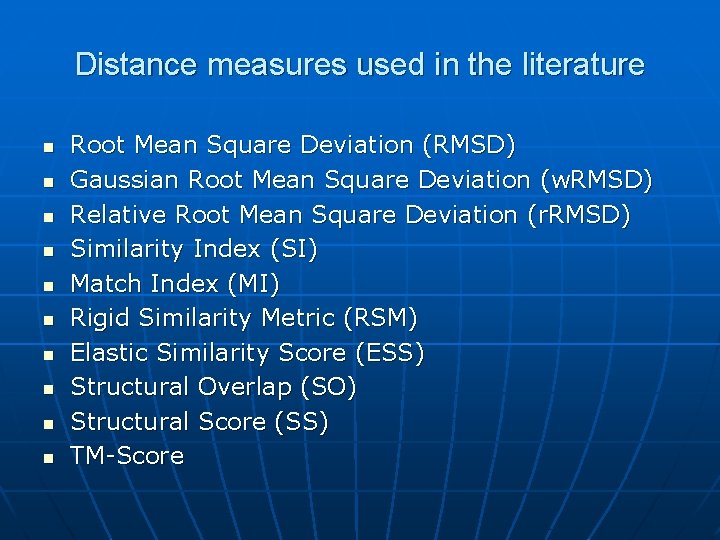
Distance measures used in the literature n n n n n Root Mean Square Deviation (RMSD) Gaussian Root Mean Square Deviation (w. RMSD) Relative Root Mean Square Deviation (r. RMSD) Similarity Index (SI) Match Index (MI) Rigid Similarity Metric (RSM) Elastic Similarity Score (ESS) Structural Overlap (SO) Structural Score (SS) TM-Score

The Program n n n PROTEUS-300, Homstrad, etc. , protein databases to choose proteins from Protein data bank as source 3 D-information, and GO Id. for functional annotations A structure alignment algorithm (Ye et al. , 2004) to align a pair of proteins’ structures, and then … Measure the “distance” or “similarity” according to each “metric” Which “metric” stands up to Go-Id similarity or protein property similarity?
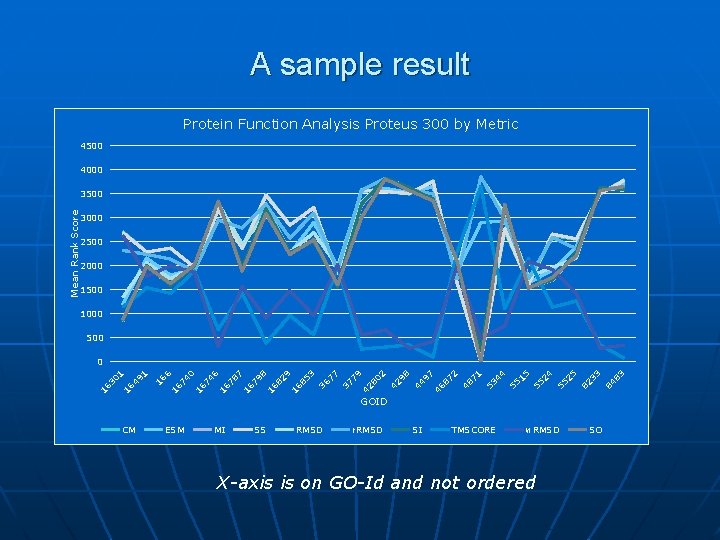
A sample result Protein Function Analysis Proteus 300 by Metric 4500 4000 Mean Rank Score 3500 3000 2500 2000 1500 1000 500 83 84 33 82 25 55 55 24 15 55 44 53 2 71 48 87 97 46 44 98 2 42 80 79 42 37 3 77 36 85 16 16 82 9 8 79 7 16 78 6 16 74 0 6 16 16 1 49 16 16 30 1 0 GOID CM ESM MI SS RMSD r. RMSD SI TMSCORE w. RMSD X-axis is on GO-Id and not ordered SO
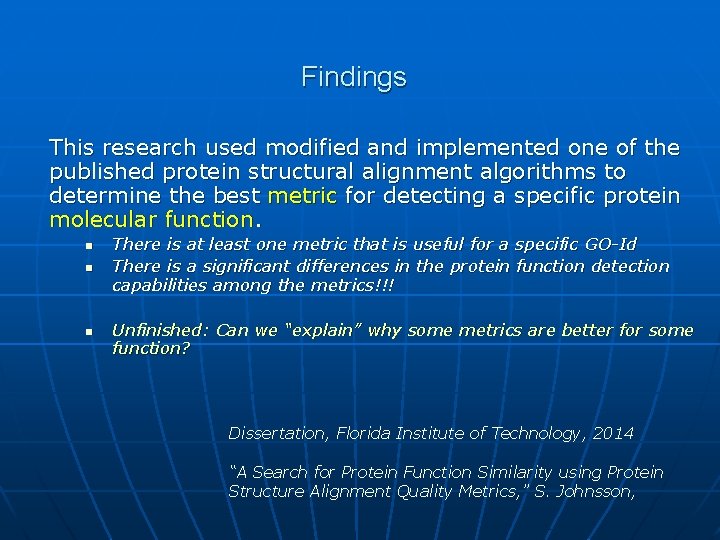
Findings This research used modified and implemented one of the published protein structural alignment algorithms to determine the best metric for detecting a specific protein molecular function. n n n There is at least one metric that is useful for a specific GO-Id There is a significant differences in the protein function detection capabilities among the metrics!!! Unfinished: Can we “explain” why some metrics are better for some function? Dissertation, Florida Institute of Technology, 2014 “A Search for Protein Function Similarity using Protein Structure Alignment Quality Metrics, ” S. Johnsson,

Project II Cell Tracking on Fluorescent Microscopy 2 D x t Debasis Mitra Rostyslav Bouchko Marit Nilsen-Hamilton 10/19/2021 D. Mitra at IBIIS, Stanford 12
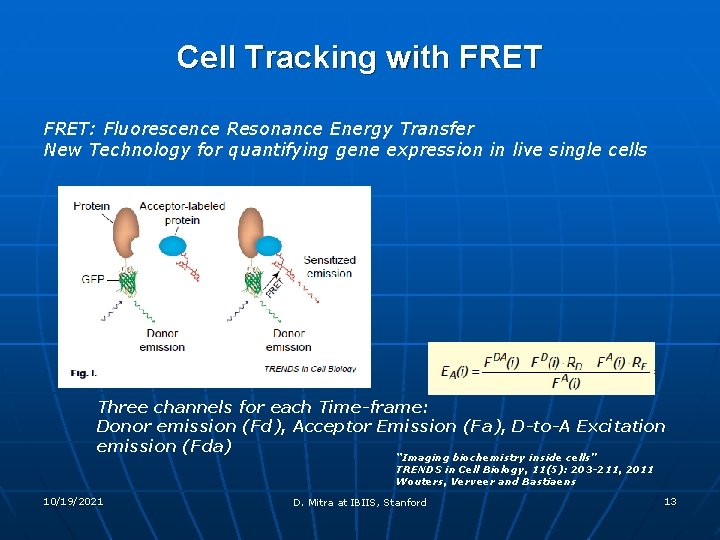
Cell Tracking with FRET: Fluorescence Resonance Energy Transfer New Technology for quantifying gene expression in live single cells Three channels for each Time-frame: Donor emission (Fd), Acceptor Emission (Fa), D-to-A Excitation emission (Fda) “Imaging biochemistry inside cells” TRENDS in Cell Biology, 11(5): 203 -211, 2011 Wouters, Verveer and Bastiaens 10/19/2021 D. Mitra at IBIIS, Stanford 13
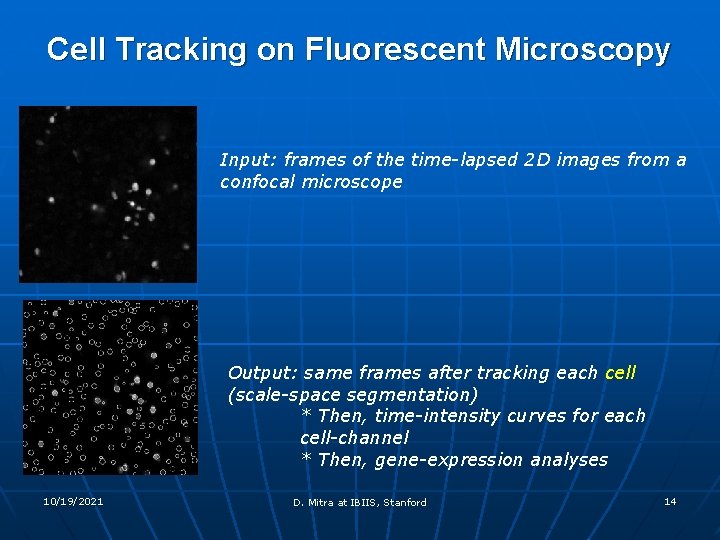
Cell Tracking on Fluorescent Microscopy Input: frames of the time-lapsed 2 D images from a confocal microscope Output: same frames after tracking each cell (scale-space segmentation) * Then, time-intensity curves for each cell-channel * Then, gene-expression analyses 10/19/2021 D. Mitra at IBIIS, Stanford 14
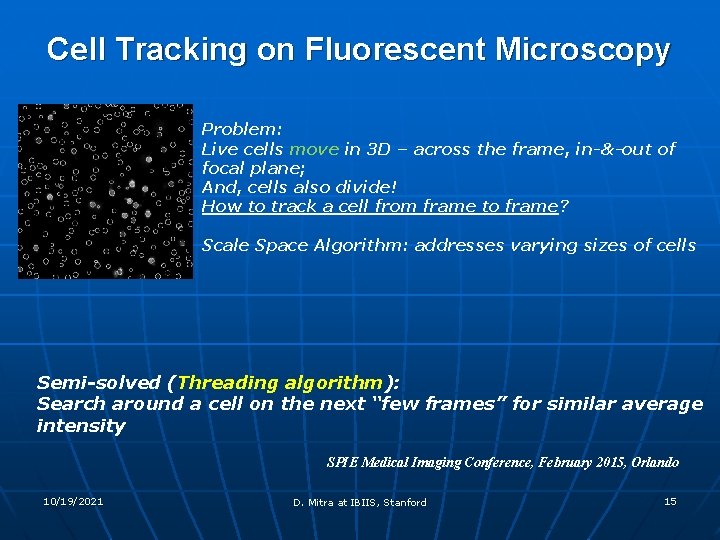
Cell Tracking on Fluorescent Microscopy Problem: Live cells move in 3 D – across the frame, in-&-out of focal plane; And, cells also divide! How to track a cell from frame to frame? Scale Space Algorithm: addresses varying sizes of cells Semi-solved (Threading algorithm): Search around a cell on the next “few frames” for similar average intensity SPIE Medical Imaging Conference, February 2015, Orlando 10/19/2021 D. Mitra at IBIIS, Stanford 15

Project III Dynamic PET* in Alzheimer’s studies: Sequence of 3 D reconstructed images * PET: Positron Emission Tomography Rostyslav Bouchko, LBL Debasis Mitra, -yours truly. William Jagust, PI @ UC Berkeley 10/19/2021 D. Mitra at IBIIS, Stanford 16
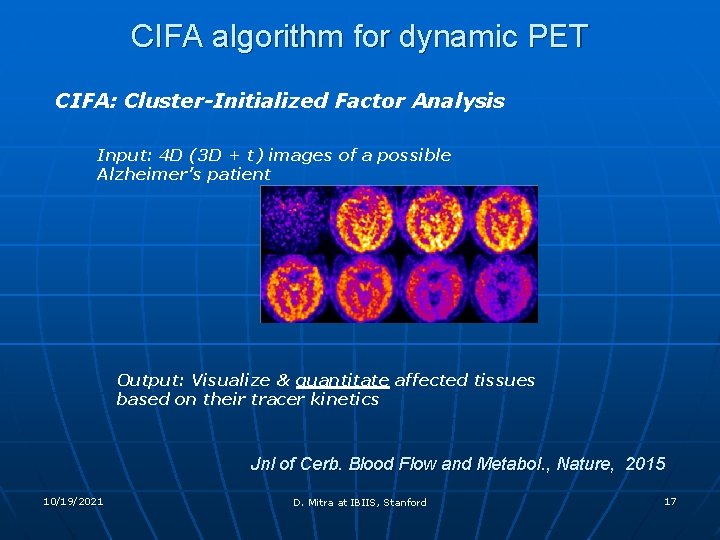
CIFA algorithm for dynamic PET CIFA: Cluster-Initialized Factor Analysis Input: 4 D (3 D + t) images of a possible Alzheimer’s patient Output: Visualize & quantitate affected tissues based on their tracer kinetics Jnl of Cerb. Blood Flow and Metabol. , Nature, 2015 10/19/2021 D. Mitra at IBIIS, Stanford 17
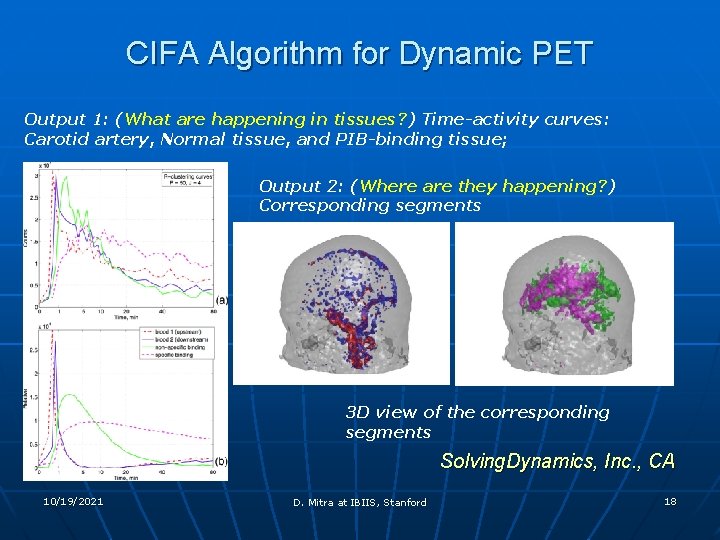
CIFA Algorithm for Dynamic PET Output 1: (What are happening in tissues? ) Time-activity curves: Carotid artery, Normal tissue, and PIB-binding tissue; Output 2: (Where are they happening? ) Corresponding segments 3 D view of the corresponding segments Solving. Dynamics, Inc. , CA 10/19/2021 D. Mitra at IBIIS, Stanford 18

Project IV Dynamic cardiac SPECT* reconstruction: From rotating camera projections * SPECT: Single Photon Emission Computed Tomography Mahmoud Abdalah, Ph. D. , Moffitt Cancer Hui Pan, Ph. D. , Tescent, China Haoran Chang, Ph. D. -candidate, FIT Debasis Mitra Rostyslav Bouchko, LBL Grant T. Gullberg, PI, LBL 10/19/2021 D. Mitra at IBIIS, Stanford 19
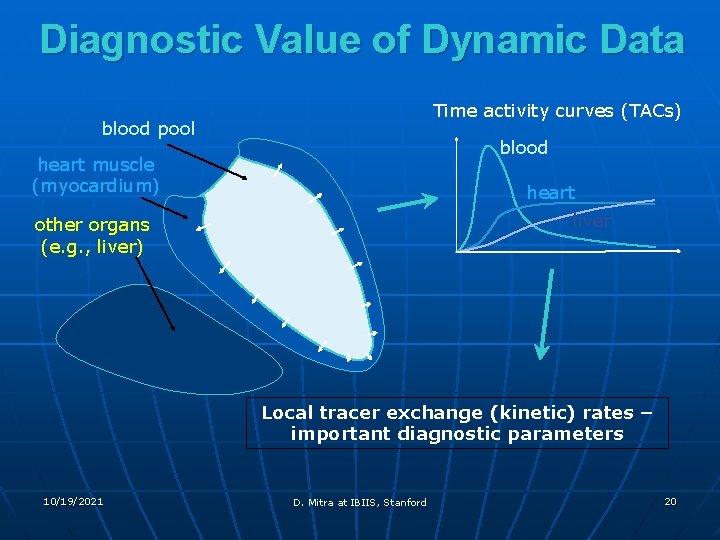
Diagnostic Value of Dynamic Data Time activity curves (TACs) blood pool blood heart muscle (myocardium) heart liver other organs (e. g. , liver) Local tracer exchange (kinetic) rates – important diagnostic parameters 10/19/2021 D. Mitra at IBIIS, Stanford 20
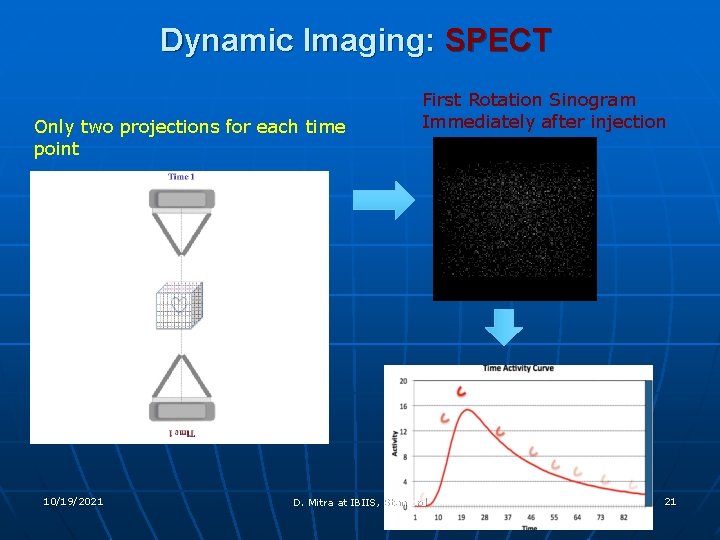
Dynamic Imaging: SPECT Only two projections for each time point 10/19/2021 First Rotation Sinogram Immediately after injection D. Mitra at IBIIS, Stanford 21
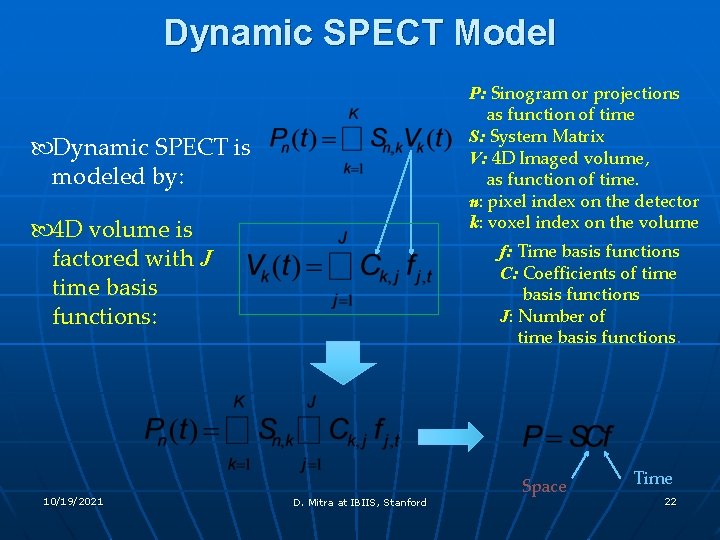
Dynamic SPECT Model P: Sinogram or projections as function of time S: System Matrix V: 4 D Imaged volume, as function of time. n: pixel index on the detector k: voxel index on the volume Dynamic SPECT is modeled by: 4 D volume is factored with J time basis functions: 10/19/2021 f: Time basis functions C: Coefficients of time basis functions J: Number of time basis functions. D. Mitra at IBIIS, Stanford Space Time 22
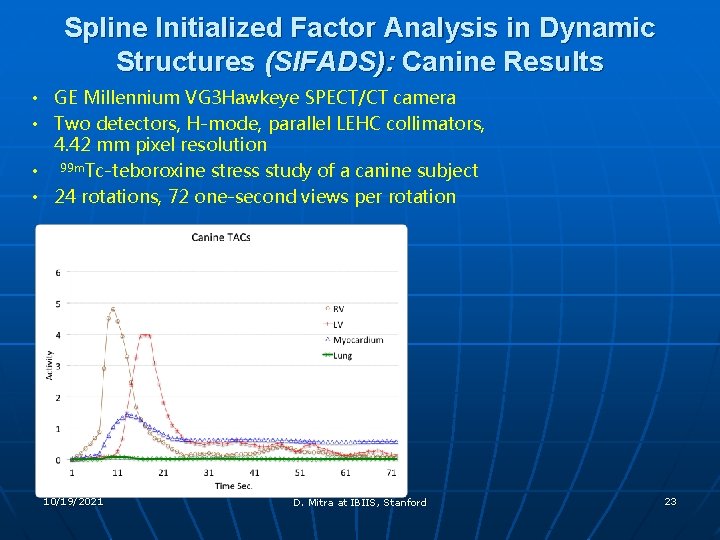
Spline Initialized Factor Analysis in Dynamic Structures (SIFADS): Canine Results • GE Millennium VG 3 Hawkeye SPECT/CT camera • Two detectors, H-mode, parallel LEHC collimators, 4. 42 mm pixel resolution • 99 m. Tc-teboroxine stress study of a canine subject • 24 rotations, 72 one-second views per rotation 10/19/2021 D. Mitra at IBIIS, Stanford 23

Original projections: Murine data imaged with pinhole camera Estimated rat TACs from the first inconsistent rotation: Reproduced projections by forward projecting dynamic reconstruction: n 10/19/2021 D. Mitra at IBIIS, Stanford s 24

Project V Breast cancer prognosis with PET-MR image (finishing) Gengbo Liu, ABD (defended) Debasis Mitra, FIT Benjamin Franc, Stanford Youngho Seo, PI @ UC San Francisco 10/19/2021 D. Mitra at IBIIS, Stanford 25
Radiomics: Image bio-markers to phenotypes (Similar to genomics) (1) Does an image has enough information content, e. g. about Estrogen Receptor, Progestoren Receptor, Triplenegative, HER 2, Tumor stage, …? (2) If yes, then what are the Radiomic biomarkers? (3) Is dual modality (PET-MR) better than single modality imaging? 10/19/2021 D. Mitra at IBIIS, Stanford 26
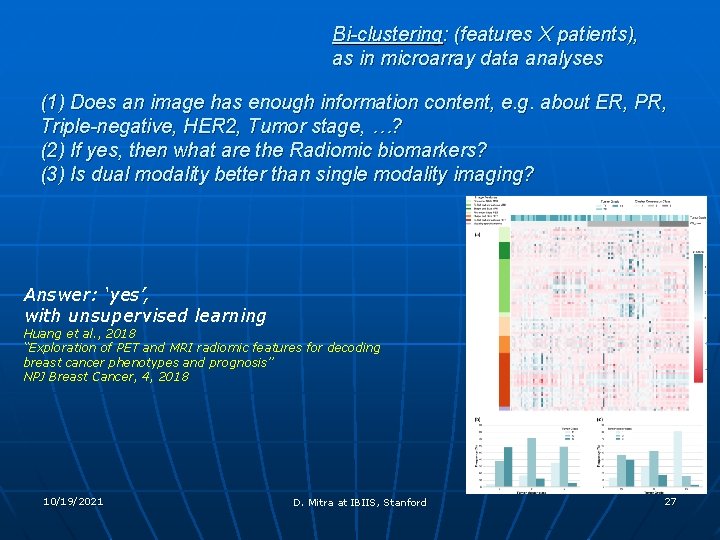
Bi-clustering: (features X patients), as in microarray data analyses (1) Does an image has enough information content, e. g. about ER, PR, Triple-negative, HER 2, Tumor stage, …? (2) If yes, then what are the Radiomic biomarkers? (3) Is dual modality better than single modality imaging? Answer: ‘yes’, with unsupervised learning Huang et al. , 2018 “Exploration of PET and MRI radiomic features for decoding breast cancer phenotypes and prognosis” NPJ Breast Cancer, 4, 2018 10/19/2021 D. Mitra at IBIIS, Stanford 27

(1) Does an image has enough information content, e. g. about ER, PR, Triple-negative, HER 2, Tumor stage, …? (2) If yes, then what are the Radiomic biomarkers? (3) Is dual modality better than single modality imaging? (1) Yes: with deep learning over radiomic features on tumors: Liu et al. , (submitted, NPJ) (2) Found (or at least, have shown a way to find) a linear combination of a subset of an exhaustive list (837) of image-statistical features (3) Dual or each of PET and MR is better for each respective item of prognosis 10/19/2021 D. Mitra at IBIIS, Stanford 28

Project VI Dynamic Contrast-enhanced Ultrasound image of Mice Colorectal Cancer (initiating) Valerie Kobrazenko, Ph. D. student Debasis Mitra, FIT Hersh Sagreiya, U. Penn Amelie Lutz & Daniel Rubin, Stanford Medicine 10/19/2021 D. Mitra at IBIIS, Stanford 29
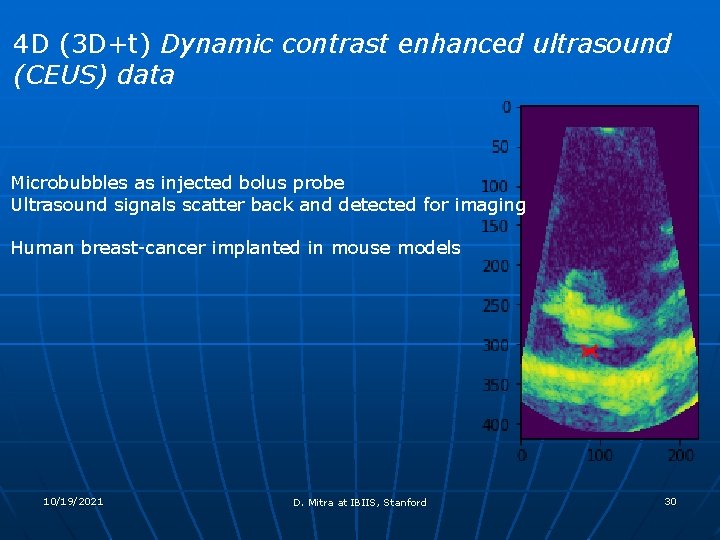
4 D (3 D+t) Dynamic contrast enhanced ultrasound (CEUS) data Microbubbles as injected bolus probe Ultrasound signals scatter back and detected for imaging Human breast-cancer implanted in mouse models 10/19/2021 D. Mitra at IBIIS, Stanford 30
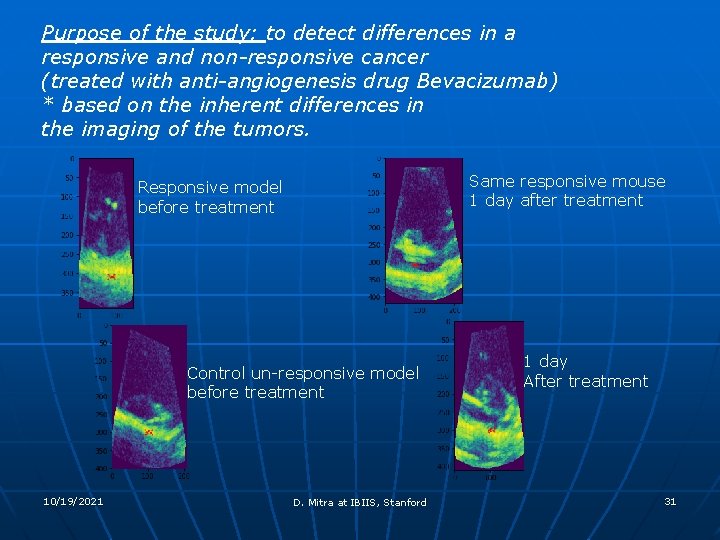
Purpose of the study: to detect differences in a responsive and non-responsive cancer (treated with anti-angiogenesis drug Bevacizumab) * based on the inherent differences in the imaging of the tumors. Same responsive mouse 1 day after treatment Responsive model before treatment Control un-responsive model before treatment 10/19/2021 D. Mitra at IBIIS, Stanford 1 day After treatment 31
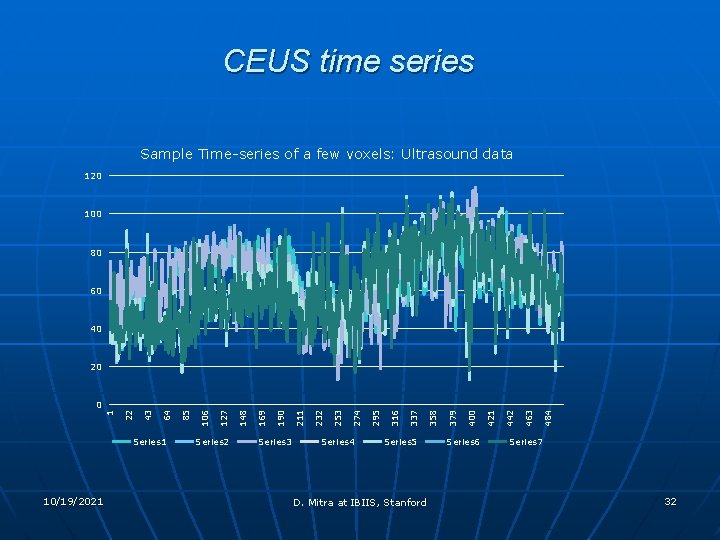
CEUS time series Sample Time-series of a few voxels: Ultrasound data 120 100 80 60 40 20 Series 1 10/19/2021 Series 2 Series 3 Series 4 Series 5 D. Mitra at IBIIS, Stanford Series 6 484 463 442 421 400 379 358 337 316 295 274 253 232 211 190 169 148 127 106 85 64 43 22 1 0 Series 7 32
Topology of time-series 10/19/2021 D. Mitra at IBIIS, Stanford 33
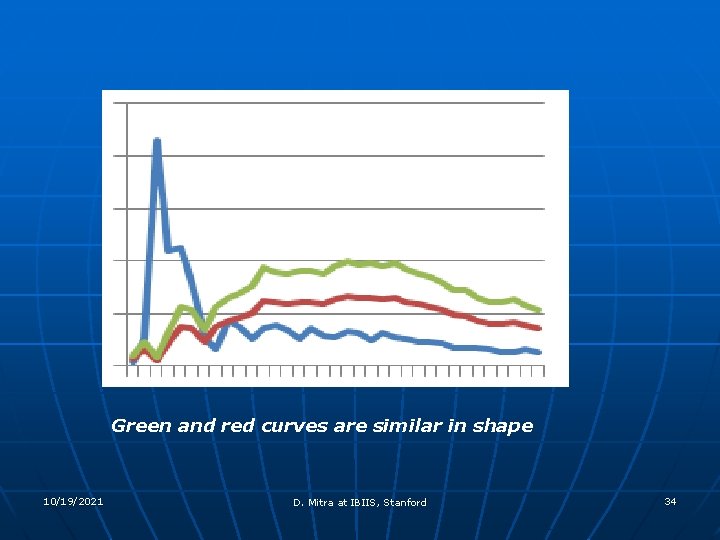
Green and red curves are similar in shape 10/19/2021 D. Mitra at IBIIS, Stanford 34
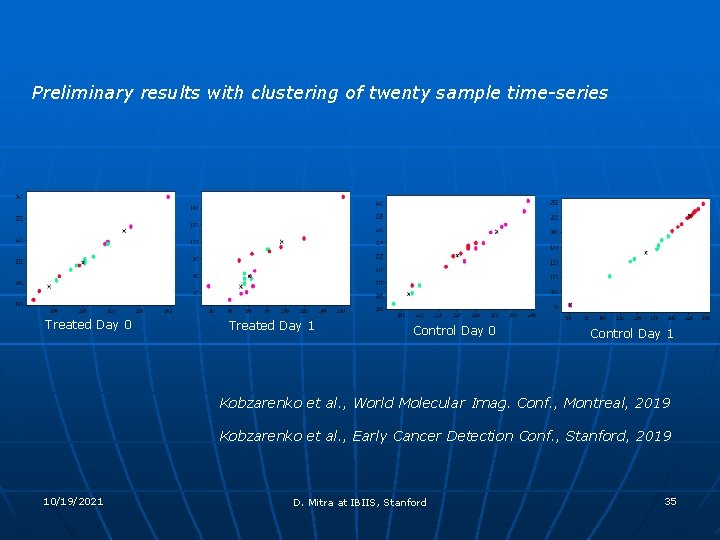
Preliminary results with clustering of twenty sample time-series Treated Day 0 Treated Day 1 Control Day 0 Control Day 1 Kobzarenko et al. , World Molecular Imag. Conf. , Montreal, 2019 Kobzarenko et al. , Early Cancer Detection Conf. , Stanford, 2019 10/19/2021 D. Mitra at IBIIS, Stanford 35
IN LIEU OF CONCLUSION (1) Dynamic data needs to be exploited in molecular imaging (2) Topological analysis provides better hypothesis formulation (3) Data science and machine learning may be keys to rich biological investigation 10/19/2021 D. Mitra at IBIIS, Stanford 36

Toda! Merci! Danke! Gracias! Thank you! Dhanyabad! Happy Diwali’ 2019!! Debasis Mitra dmitra@cs. fit. edu 10/19/2021 D. Mitra at IBIIS, Stanford 37
- Slides: 37