A Computer Aided Detection System For Digital Mammograms
















- Slides: 30

A Computer Aided Detection System For Digital Mammograms Based on Radial Basis Functions and Feature Extraction Techniques By Mohammed Jirari Shanghai, China Sept 3 rd, 2005

Why This Project? • Breast Cancer is the most common cancer and is the second leading cause of cancer deaths • Mammographic screening reduces the mortality of breast cancer • But, mammography has low positive predictive value PPV (only 35% have malignancies) • Goal of Computer Aided Detection CAD is to provide a second reading, hence reducing the false positive rate

Basic Components of the System • Preprocessing – Cropping – Enhancement (Histogram Equalization) • • • Feature extraction Normalization Training Testing ROC Analysis

What is a Mammogram? • A Mammogram is an x-ray image of the breast. Mammography is the procedure used to generate a mammogram • The equipment used to obtain a mammogram, however, is very different from that used to perform an x-ray of chest or bones

Mammograms (cont. ) • In order to get a good image, the breast must also be flattened or compressed • In a standard examination, two images of each breast are taken: one from the top and one from the side

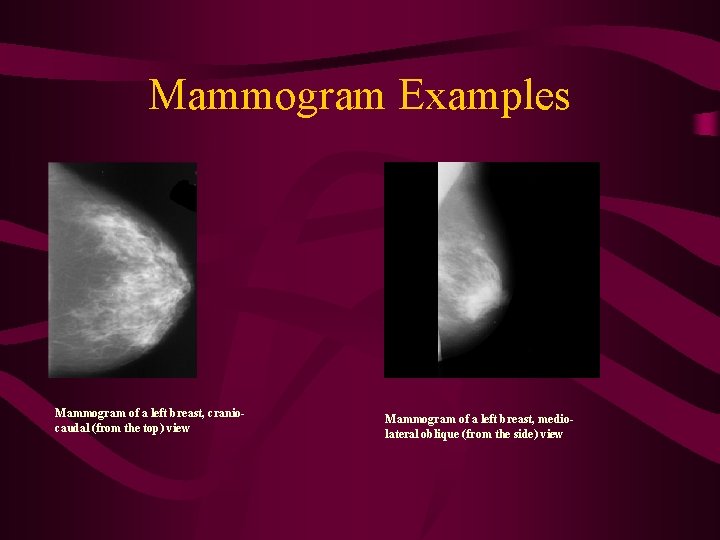
Mammogram Examples Mammogram of a left breast, craniocaudal (from the top) view Mammogram of a left breast, mediolateral oblique (from the side) view

Purpose of CAD • Mammography is the most reliable method in early detection of breast cancer • But, due to the high number of mammograms to be read, the accuracy rate tends to decrease • Double reading of mammograms has been proven to increase the accuracy, but at high cost • CAD can assist the medical staff to achieve high efficiency and effectiveness • The physician/radiologist makes the call not CAD

Proposed Method • The proposed method will assist the physician by providing a second opinion on reading the mammogram, by pointing out an area (if one exists) delimited by its center coordinates and its radius • If the two readings are similar, no more work is to be done • If they are different, the radiologist will take a second look to make the final diagnosis

Data Used • The dataset used is the Mammographic Image Analysis Society (MIAS) MINIMIAS database containing Medio. Lateral Oblique (MLO) views for each breast for 161 patients for a total of 322 images Each image is: 1024 pixels X 1024 pixels

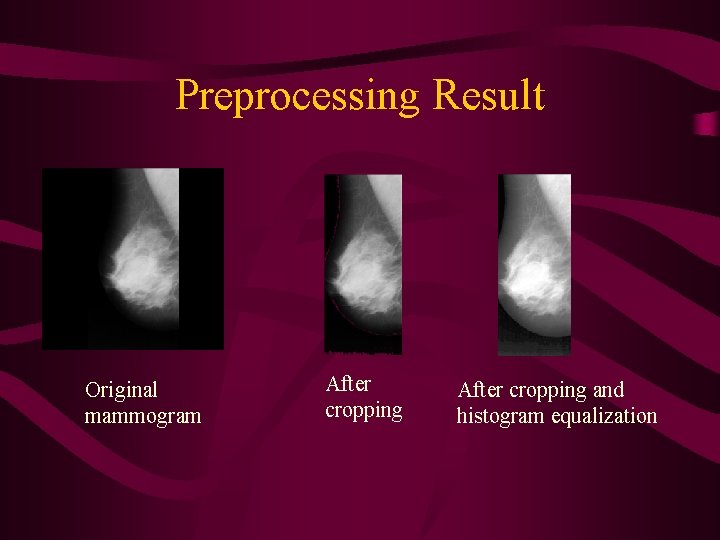
Preprocessing • Cropping: cuts the black parts of the image (almost 50%) based on a threshold • Enhancement: Histogram equalization to accentuate the features to be extracted by increasing the dynamic range of gray levels

Preprocessing Result Original mammogram After cropping and histogram equalization

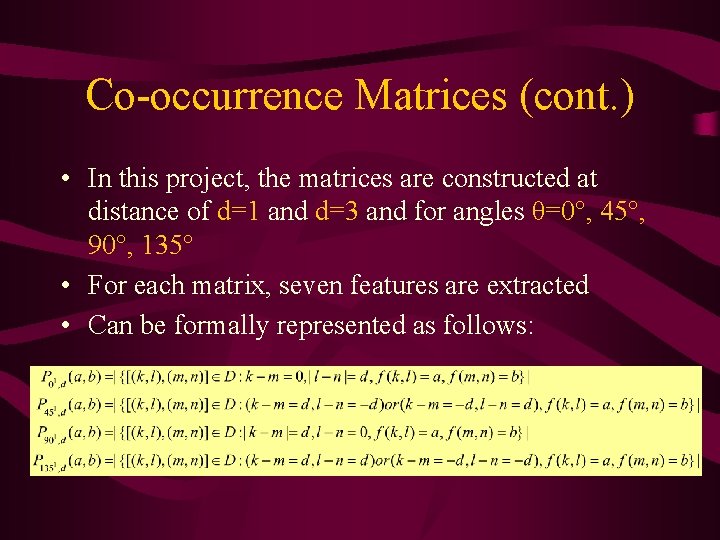
Co-occurrence Matrices to Calculate Features • The joint probability of occurrence of gray level a and b for two pixels with a defined spatial relationship in an image • The spatial relationship is defined in terms of distance d angle θ • From these matrices, a variety of features may be extracted

Co-occurrence Matrices (cont. ) • In this project, the matrices are constructed at distance of d=1 and d=3 and for angles θ=0°, 45°, 90°, 135° • For each matrix, seven features are extracted • Can be formally represented as follows:

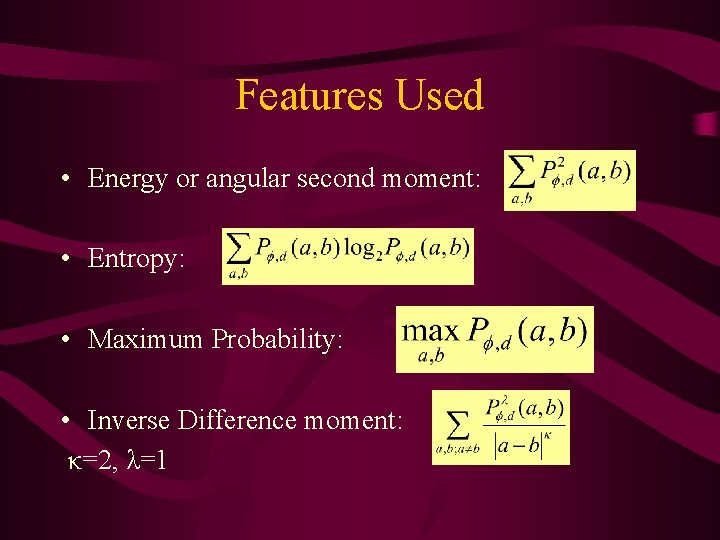
Features Used • Energy or angular second moment: • Entropy: • Maximum Probability: • Inverse Difference moment: κ=2, λ=1

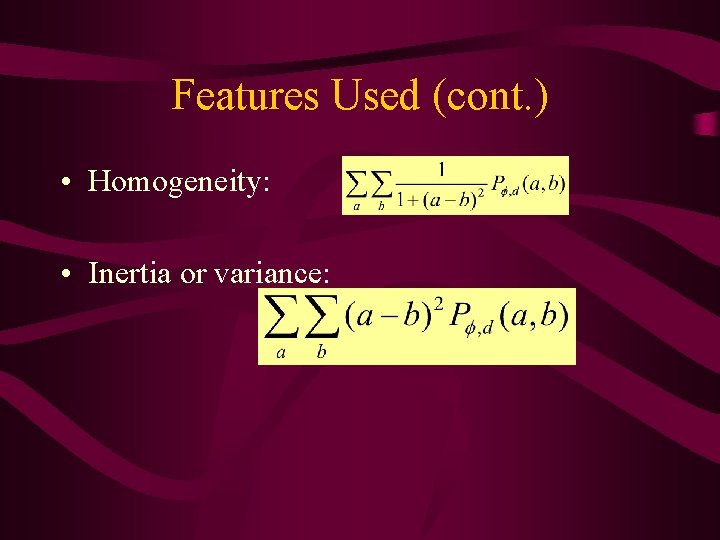
Features Used (cont. ) • Homogeneity: • Inertia or variance:

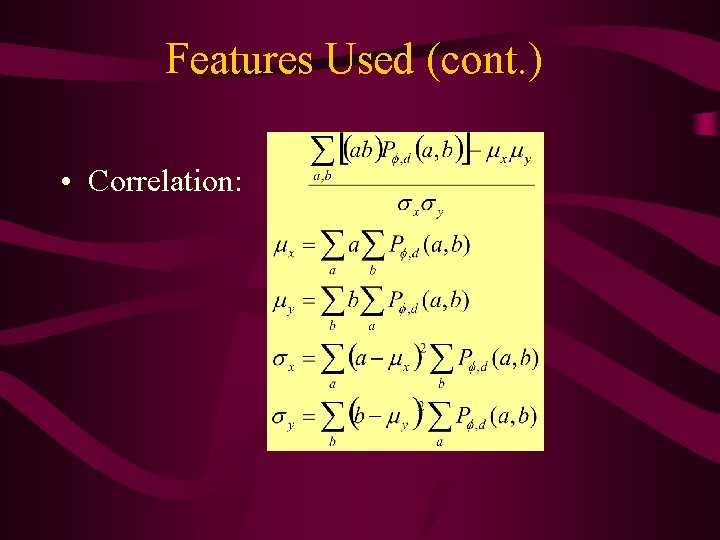
Features Used (cont. ) • Correlation:

Feature Extraction • Calculate the co-occurrence matrices at distance d=1 and d=3 • The angles used are θ=0°, 45°, 90°, 135° with the fifth matrix being the mean of the 4 directions • The co-occurrence matrices and seven statistical features are computed

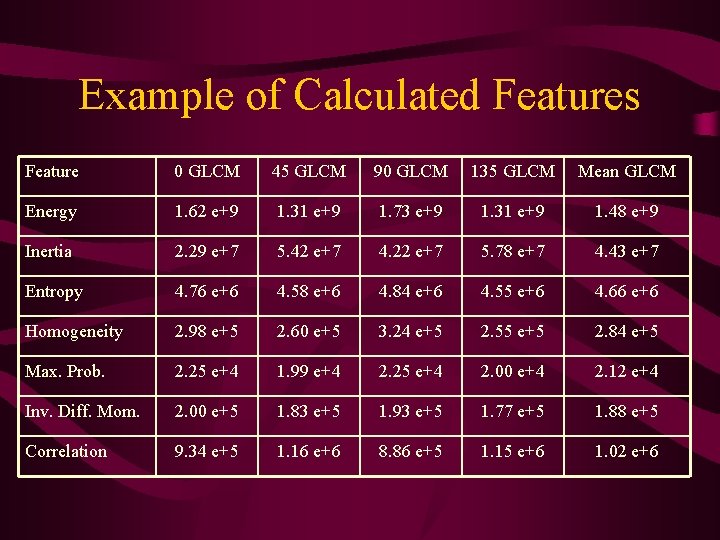
Example of Calculated Features Feature 0 GLCM 45 GLCM 90 GLCM 135 GLCM Mean GLCM Energy 1. 62 e+9 1. 31 e+9 1. 73 e+9 1. 31 e+9 1. 48 e+9 Inertia 2. 29 e+7 5. 42 e+7 4. 22 e+7 5. 78 e+7 4. 43 e+7 Entropy 4. 76 e+6 4. 58 e+6 4. 84 e+6 4. 55 e+6 4. 66 e+6 Homogeneity 2. 98 e+5 2. 60 e+5 3. 24 e+5 2. 55 e+5 2. 84 e+5 Max. Prob. 2. 25 e+4 1. 99 e+4 2. 25 e+4 2. 00 e+4 2. 12 e+4 Inv. Diff. Mom. 2. 00 e+5 1. 83 e+5 1. 93 e+5 1. 77 e+5 1. 88 e+5 Correlation 9. 34 e+5 1. 16 e+6 8. 86 e+5 1. 15 e+6 1. 02 e+6

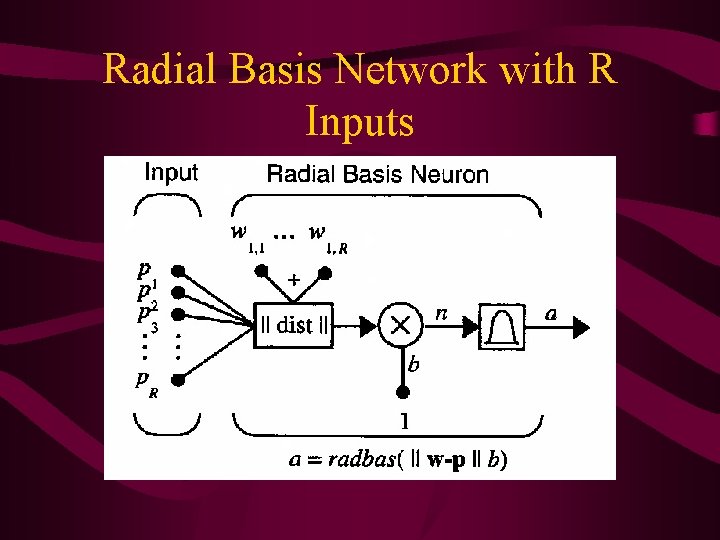
Radial Basis Network Used • Radial basis networks may require more neurons than standard feed-forward backpropagation (FFBP) networks • BUT, can be designed in a fraction of the time to train FFBP • Work best with many training vectors

Radial Basis Network with R Inputs

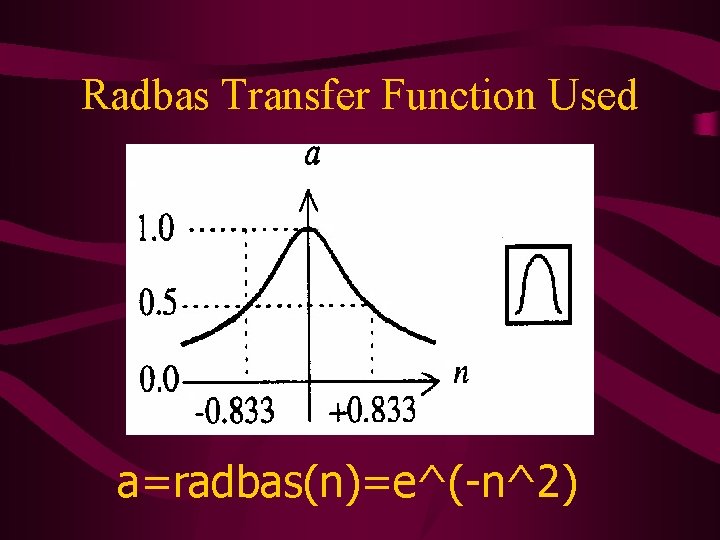
Radbas Transfer Function Used a=radbas(n)=e^(-n^2)

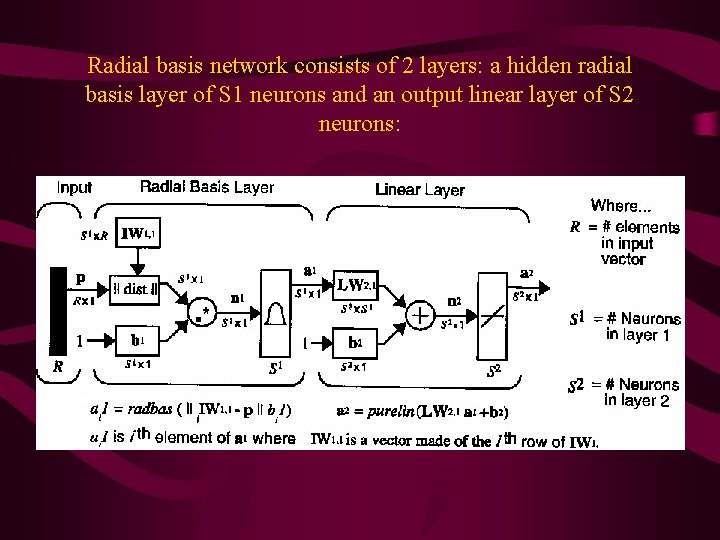
Radial basis network consists of 2 layers: a hidden radial basis layer of S 1 neurons and an output linear layer of S 2 neurons:

Training • After normalizing the data, training begins • The first training set was made up of 212 mammograms with 81 abnormal ones, with features calculated at distances d=1 and d=3 • The second training set was made up of 163 mammograms with 81 abnormal ones, with features calculated at distances d=1 and d=3

Testing • A mammogram is presented to the trained network and the output is a suspicious area denoted by its center’s x and y coordinates and its radius. If the mammogram is considered to be normal then zeros are returned for the coordinates and radius • The radiologist can then review his/her original assessment of the patient if some areas uncovered by the network were not originally looked at closely • The whole database is tested and the accuracy is calculated • The smaller dataset performed better than the larger one, and using d=3 leads to better results than d=1

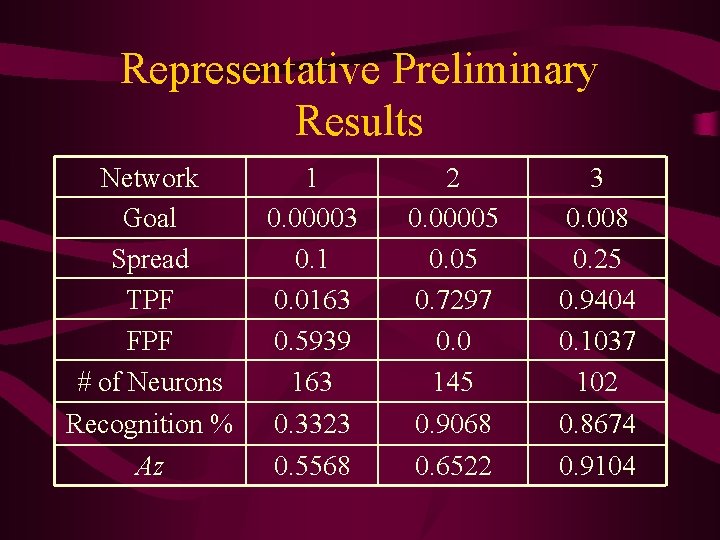
Results • • • 2 training datasets: 163 and 212 2 distance measures: 1 and 3 3 spreads: 0. 1, 0. 25, and 0. 05 3 goals: 0. 00003, 0. 008, 0. 00005 For 12 possible combinations The NN was sensitive to the unbalanced data collection that contained about 70 -30 split in the larger training set. Therefore the smaller dataset was preferred • Achieving a high recognition % is not that appealing if the TPF is small

Representative Preliminary Results Network Goal Spread TPF FPF # of Neurons 1 0. 00003 0. 1 0. 0163 0. 5939 163 2 0. 00005 0. 7297 0. 0 145 3 0. 008 0. 25 0. 9404 0. 1037 102 Recognition % 0. 3323 0. 9068 0. 8674 Az 0. 5568 0. 6522 0. 9104

Future work • Use more features like standard deviation, skewness, and kurtosis • Which feature(s) have the most impact: * Rank the features from best to worst (single input to NN) * Select most significant feature(s) by using leave one out method • Determine whether the area is benign or malignant by adding the severity of the abnormality to the training

Future work (cont. ) • Try and reduce False Negatives on the basis of region characteristics size, difference in homogeneity and entropy • Use larger database that contains both MLO and CC to train/learn, since most commercial CADs use hundreds of thousands of mammograms to try and recognize foreign samples

Thank you

Questions