A Comparative Study of Multiple Reaction Monitoring MRM
- Slides: 1

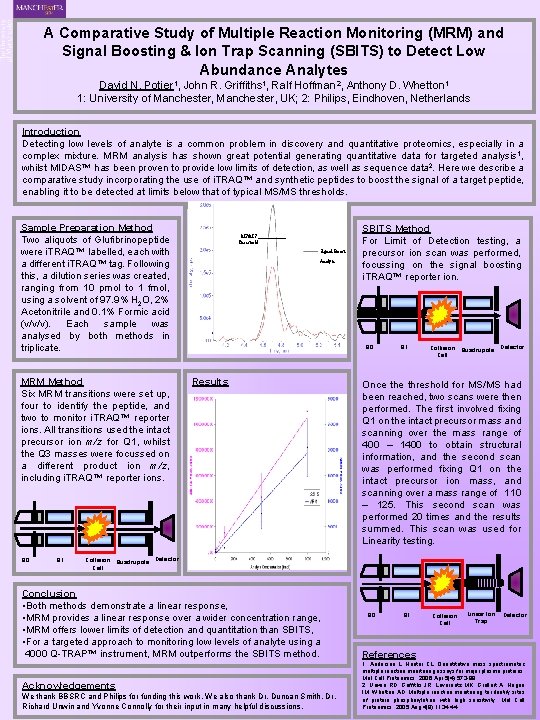
A Comparative Study of Multiple Reaction Monitoring (MRM) and Signal Boosting & Ion Trap Scanning (SBITS) to Detect Low Abundance Analytes David N. Potier 1, John R. Griffiths 1, Ralf Hoffman 2, Anthony D. Whetton 1 1: University of Manchester, UK; 2: Philips, Eindhoven, Netherlands Introduction Detecting low levels of analyte is a common problem in discovery and quantitative proteomics, especially in a complex mixture. MRM analysis has shown great potential generating quantitative data for targeted analysis 1, whilst MIDAS™ has been proven to provide low limits of detection, as well as sequence data 2. Here we describe a comparative study incorporating the use of i. TRAQ™ and synthetic peptides to boost the signal of a target peptide, enabling it to be detected at limits below that of typical MS/MS thresholds. Sample Preparation Method Two aliquots of Glufibrinopeptide were i. TRAQ™ labelled, each with a different i. TRAQ™ tag. Following this, a dilution series was created, ranging from 10 pmol to 1 fmol, using a solvent of 97. 9% H 2 O, 2% Acetonitrile and 0. 1% Formic acid (v/v/v). Each sample was analysed by both methods in triplicate. MRM Method Six MRM transitions were set up, four to identify the peptide, and two to monitor i. TRAQ™ reporter ions. All transitions used the intact precursor ion m/z for Q 1, whilst the Q 3 masses were focussed on a different product ion m/z, including i. TRAQ™ reporter ions. Q 0 Q 1 MS/MS Threshold Signal Boost Analyte SBITS Method For Limit of Detection testing, a precursor ion scan was performed, focussing on the signal boosting i. TRAQ™ reporter ion. Q 0 Results Q 1 Collision Quadrupole Cell Detector Once threshold for MS/MS had been reached, two scans were then performed. The first involved fixing Q 1 on the intact precursor mass and scanning over the mass range of 400 – 1400 to obtain structural information, and the second scan was performed fixing Q 1 on the intact precursor ion mass, and scanning over a mass range of 110 – 125. This second scan was performed 20 times and the results summed. This scan was used for Linearity testing. Collision Quadrupole Detector Cell Conclusion • Both methods demonstrate a linear response, • MRM provides a linear response over a wider concentration range, • MRM offers lower limits of detection and quantitation than SBITS, • For a targeted approach to monitoring low levels of analyte using a 4000 Q-TRAP™ instrument, MRM outperforms the SBITS method. Acknowledgements We thank BBSRC and Philips for funding this work. We also thank Dr. Duncan Smith, Dr. Richard Unwin and Yvonne Connolly for their input in many helpful discussions. Q 0 Q 1 Collision Cell Linear Ion Trap Detector References 1: Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006 Apr; 5(4): 573 -88. 2: Unwin RD, Griffiths JR, Leverentz MK, Grallert A, Hagan IM, Whetton AD. Multiple reaction monitoring to identify sites of protein phosphorylation with high sensitivity. Mol Cell Proteomics. 2005 Aug; 4(8): 1134 -44.

