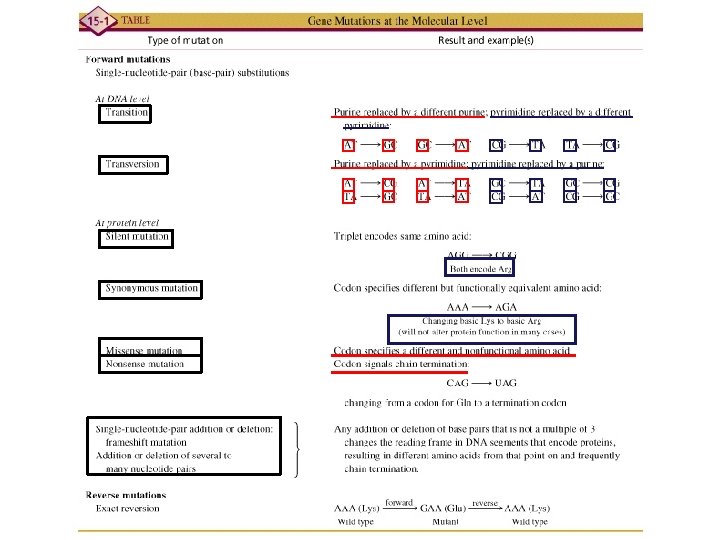
a a mutation forward mutation a a reverse















- Slides: 57

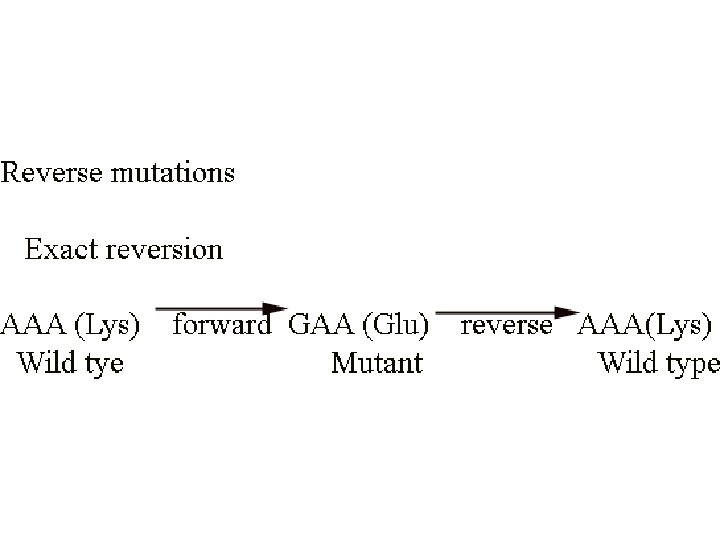
a+ --> a- mutation (forward mutation) a- --> a+ reverse mutation (reversion)

Presence of pink pigment Four-o’clock plants Incomplete dominance the term used to describe the general case in which the phenotype of a heterozygote is intermediate between those of the two homozygotes, on some quantitative scale of measurement +/-

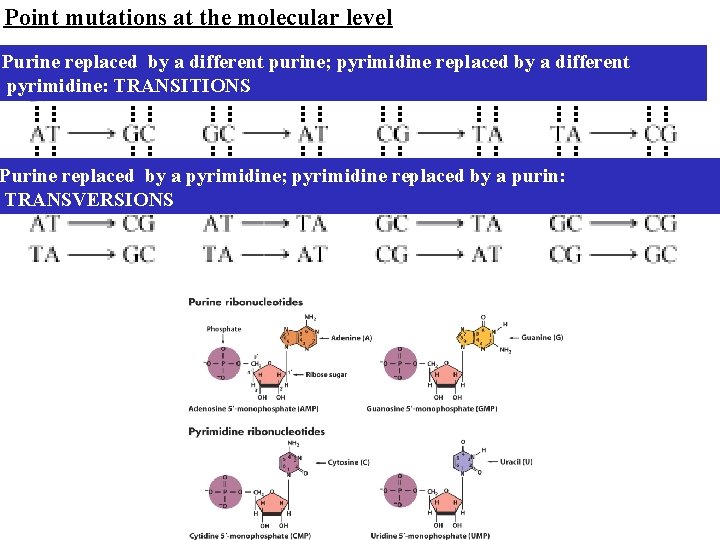
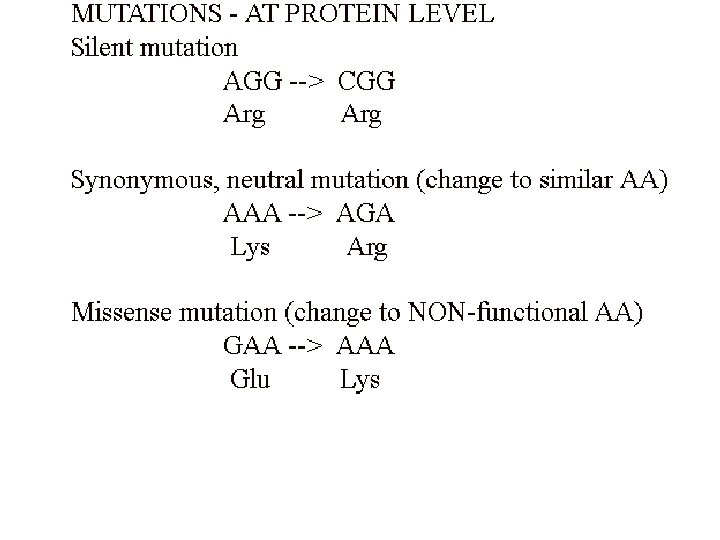
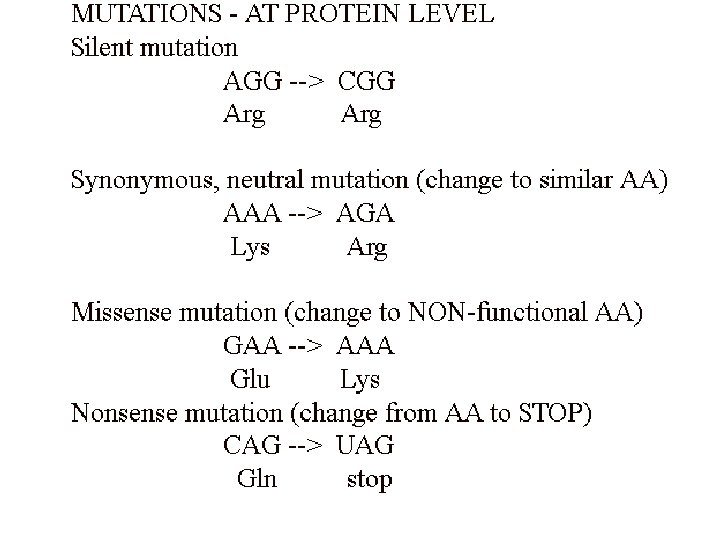
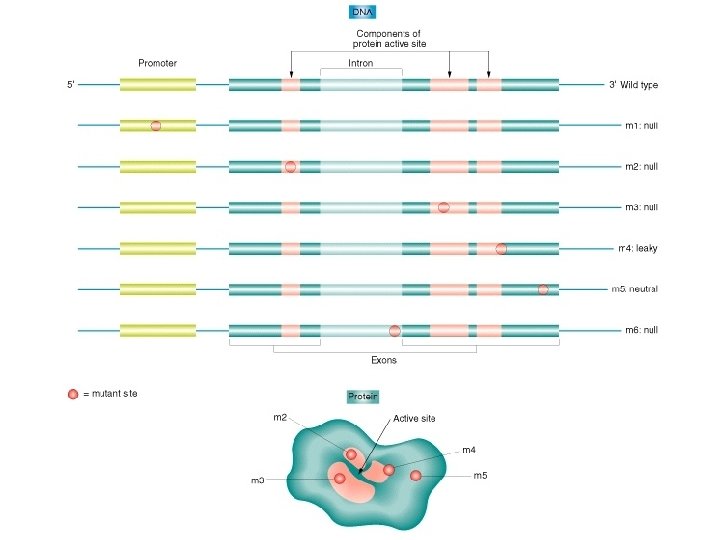
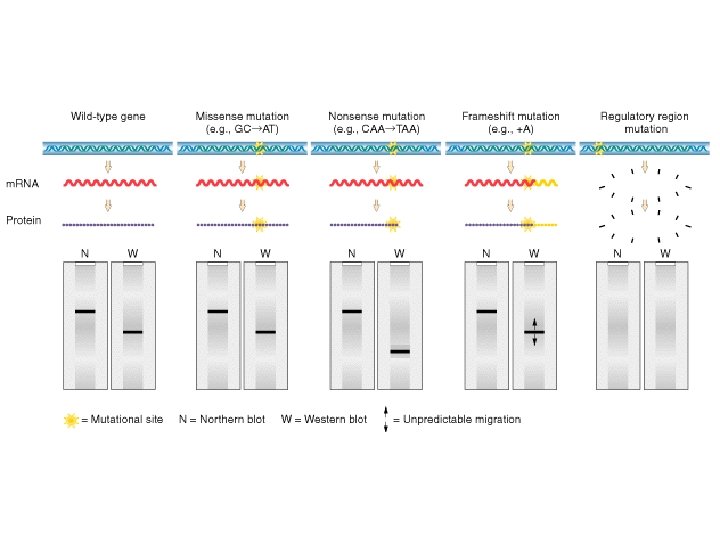
Point mutations at the molecular level Purine replaced by a different purine; pyrimidine replaced by a different pyrimidine: TRANSITIONS Purine replaced by a pyrimidine; pyrimidine replaced by a purin: TRANSVERSIONS



Animation ed. 9: 9. 2&9. 17 TRANSLATION


Animation ed. 9: 9. 2&9. 17 TRANSLATION





Now, insertions and deletions of base pairs:


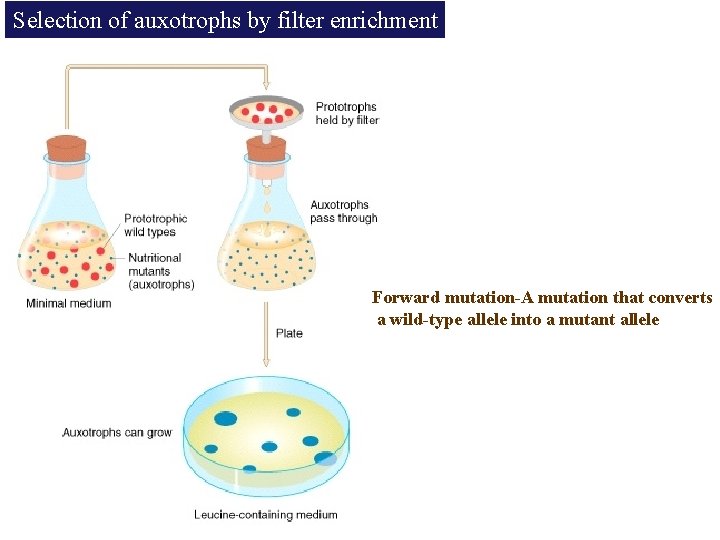
Selection of auxotrophs by filter enrichment Forward mutation-A mutation that converts a wild-type allele into a mutant allele

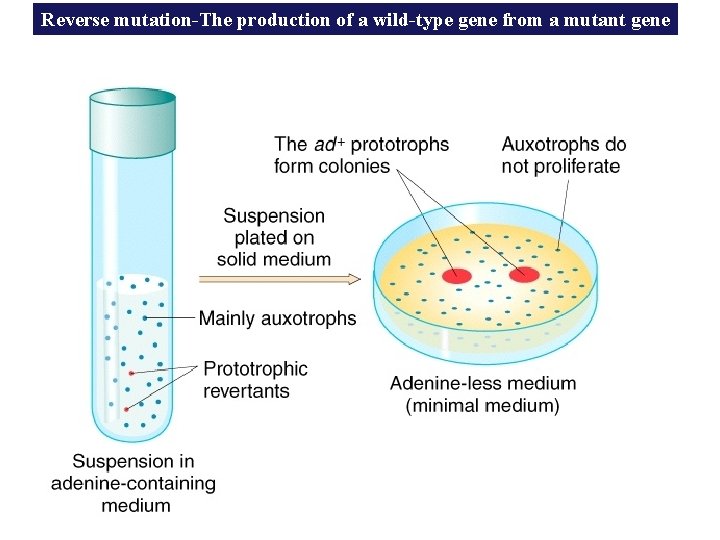
Reverse mutation-The production of a wild-type gene from a mutant gene


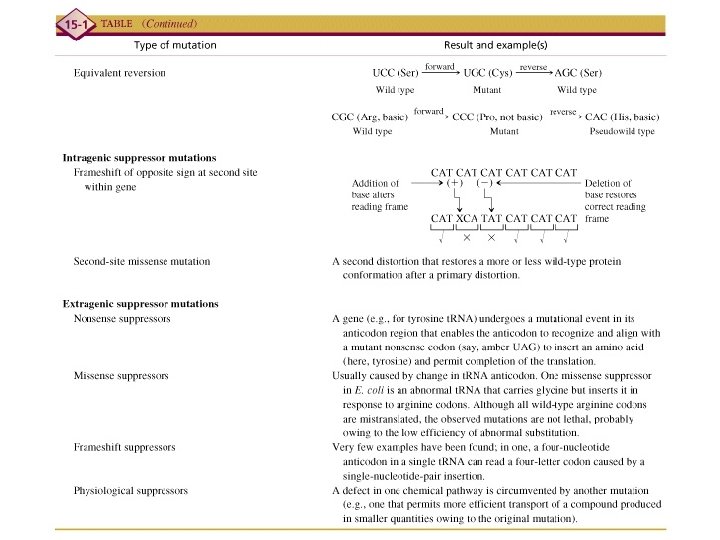
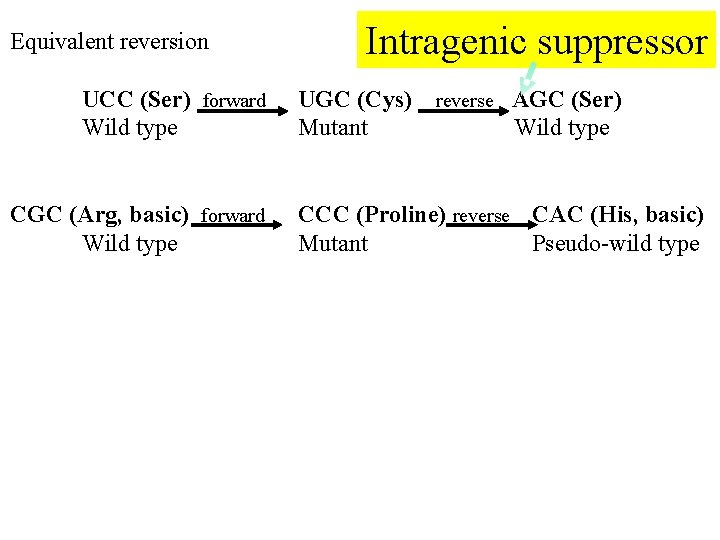
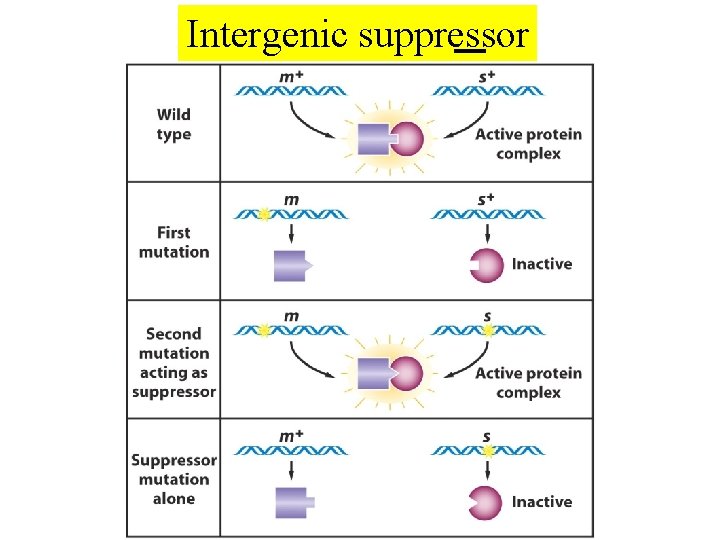
Equivalent reversion UCC (Ser) forward Wild type CGC (Arg, basic) forward Wild type Intragenic suppressor UGC (Cys) Mutant reverse AGC (Ser) Wild type CCC (Proline) reverse CAC (His, basic) Mutant Pseudo-wild type

Intragenic suppressor

Intergenic suppressor

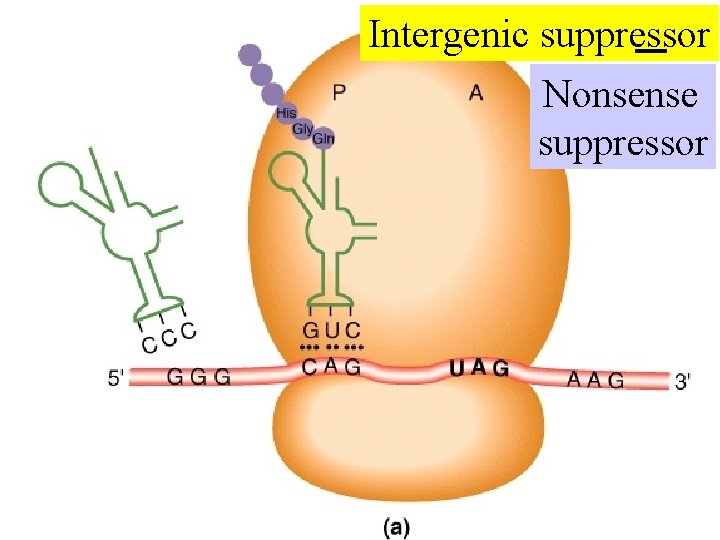
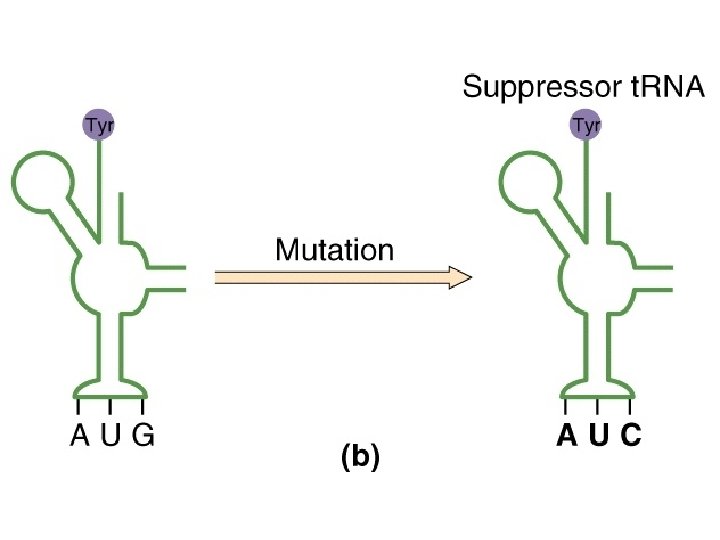
Intergenic suppressor Nonsense suppressor


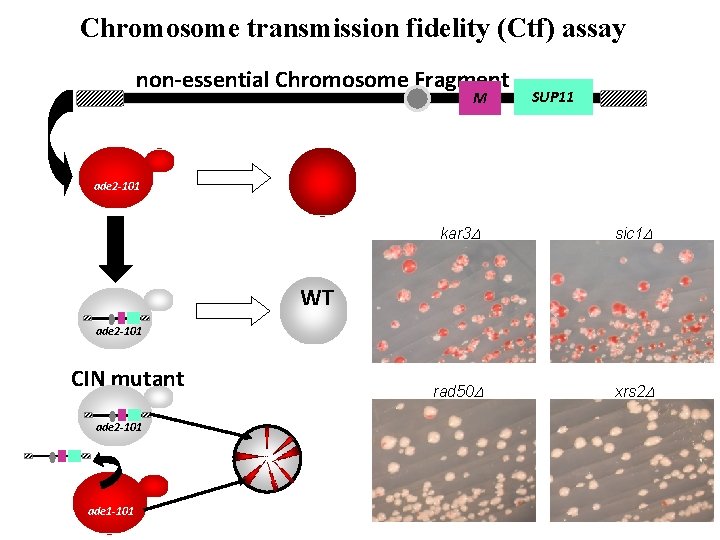
Chromosome transmission fidelity (Ctf) assay non-essential Chromosome Fragment M SUP 11 ade 2 -101 kar 3 D sic 1 D rad 50 D xrs 2 D WT ade 2 -101 CIN mutant ade 2 -101 ade 1 -101

Animation ed 9: 9. 19 a Nonsense mutation

Animation ed 9: 9. 19 b Nonsense suppressor

Animation 9. 19 c Nonsense suppression rodns and suppressor-t. RNA together give WT phenotype

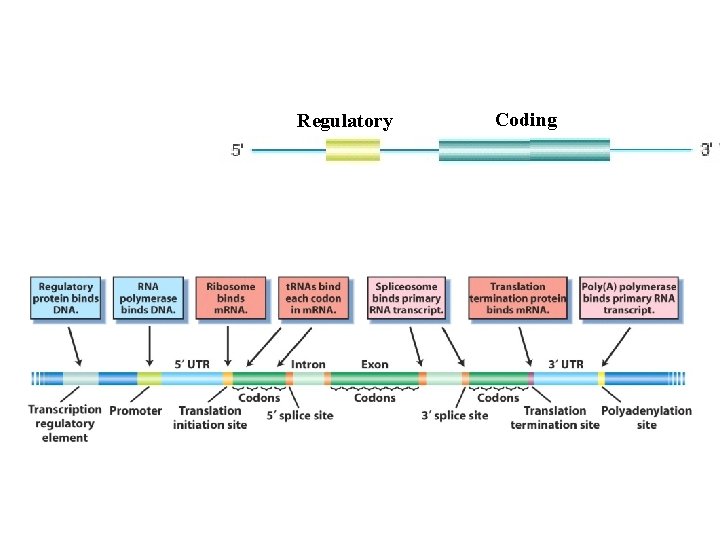
Regulatory Coding



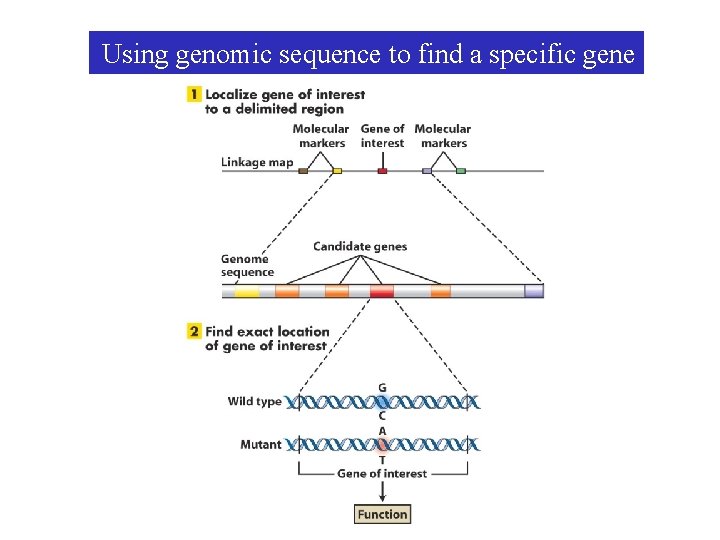
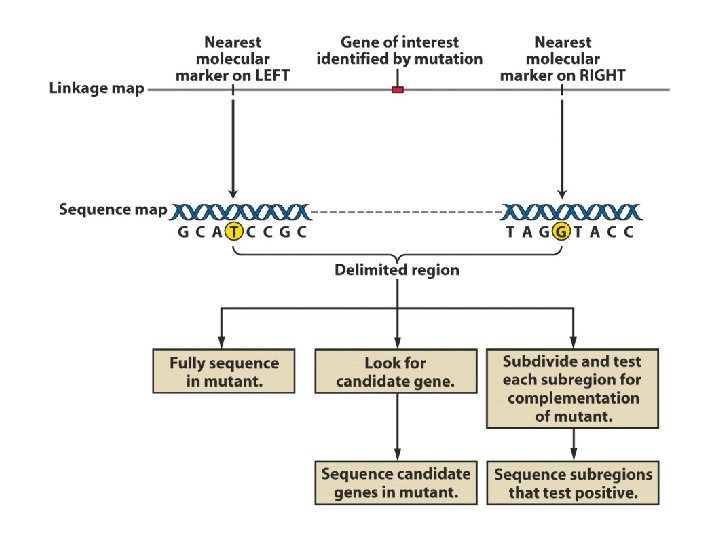
Using genomic sequence to find a specific gene

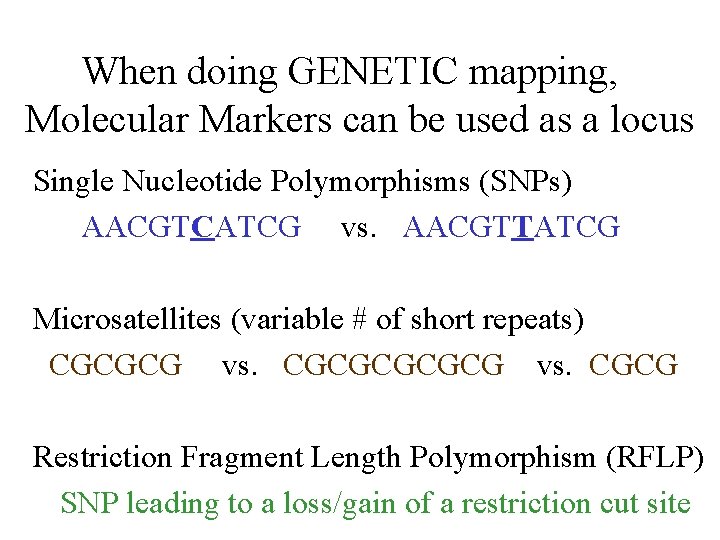
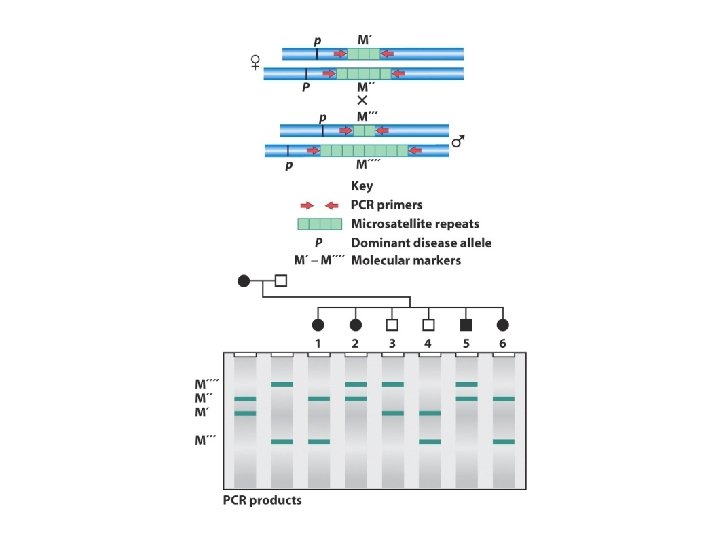
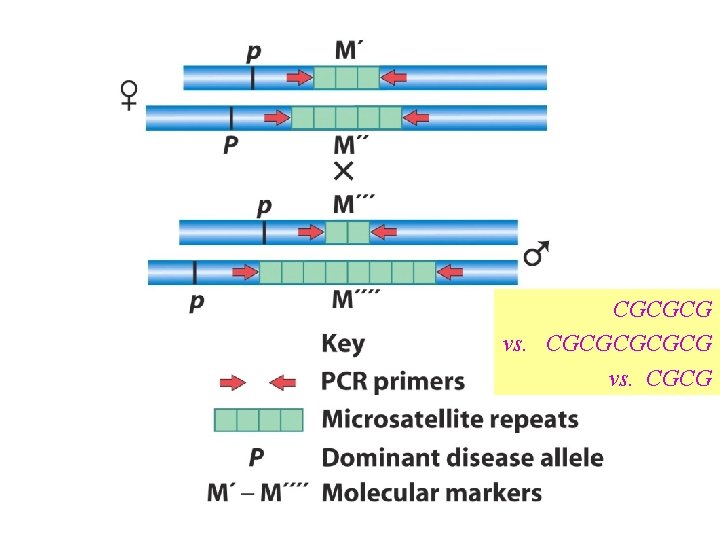
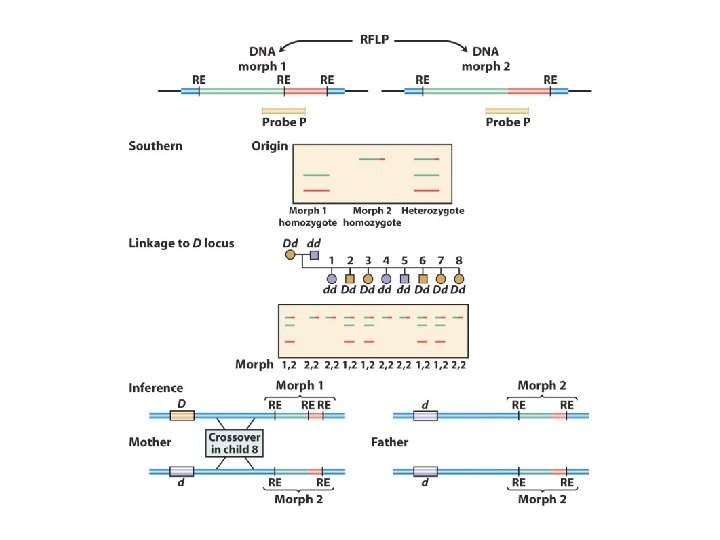
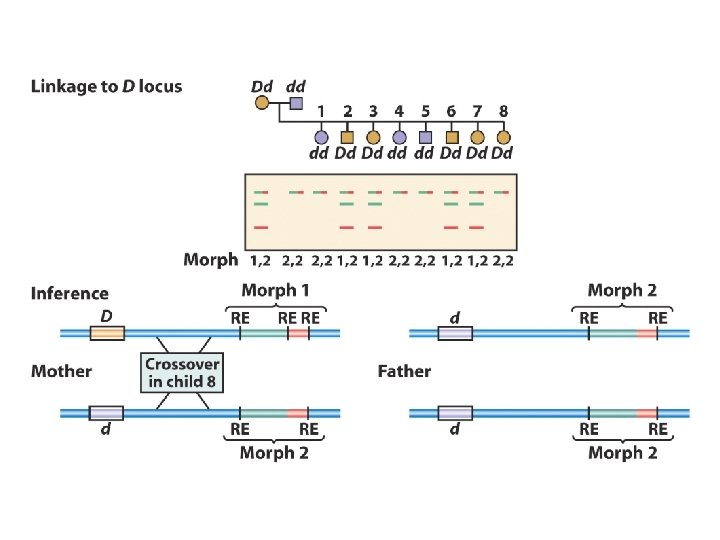
When doing GENETIC mapping, Molecular Markers can be used as a locus Single Nucleotide Polymorphisms (SNPs) AACGTCATCG vs. AACGTTATCG Microsatellites (variable # of short repeats) CGCGCG vs. CGCG Restriction Fragment Length Polymorphism (RFLP) SNP leading to a loss/gain of a restriction cut site

When doing GENETIC mapping, Molecular Markers can be used as a locus Almost all SNPs, Microsatellites, etc. are SILENT, and there are millions of them They are mile-markers, not destinations! , אבני דרך ! ולא יעדים

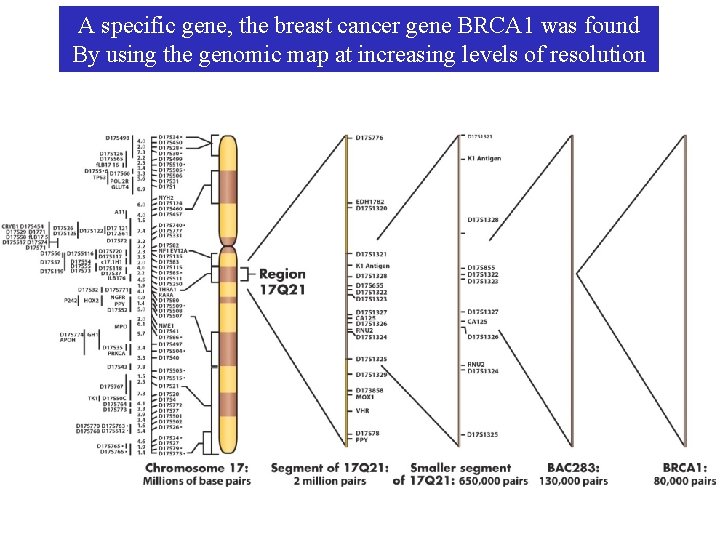
A specific gene, the breast cancer gene BRCA 1 was found By using the genomic map at increasing levels of resolution

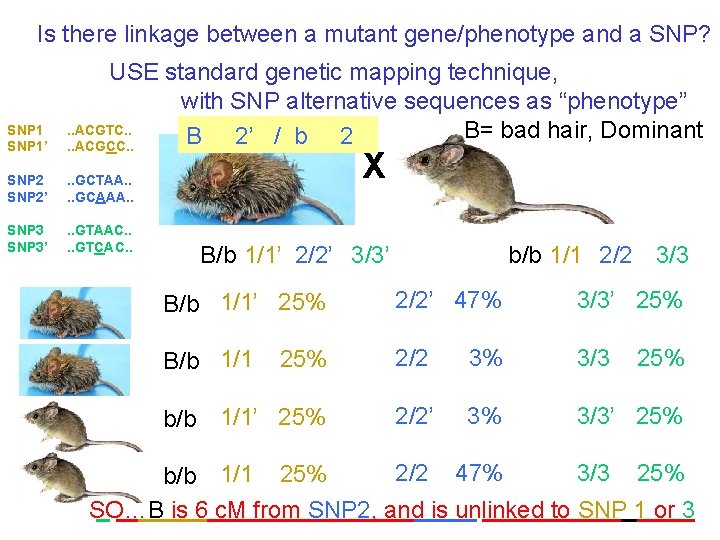
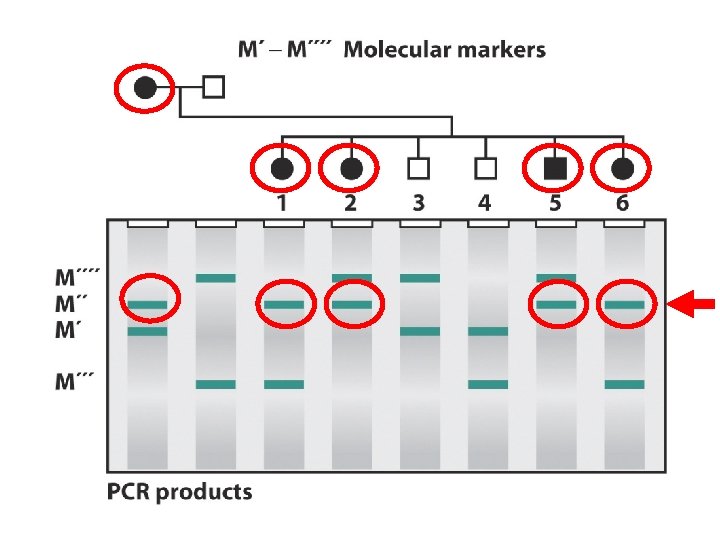
Is there linkage between a mutant gene/phenotype and a SNP? SNP 1’ USE standard genetic mapping technique, with SNP alternative sequences as “phenotype”. . ACGTC. . B= bad hair, Dominant B 2’ / b 2. . ACGCC. . SNP 2’ . . GCTAA. . GCAAA. . SNP 3’ . . GTAAC. . GTCAC. . X B/b 1/1’ 2/2’ 3/3’ b/b 1/1 2/2 3/3 B/b 1/1’ 25% 2/2’ 47% 3/3’ 25% B/b 1/1 25% 2/2 3% 3/3 b/b 1/1’ 25% 2/2’ 3% 3/3’ 25% 2/2 47% 3/3 25% b/b 1/1 25% SO…B is 6 c. M from SNP 2, and is unlinked to SNP 1 or 3

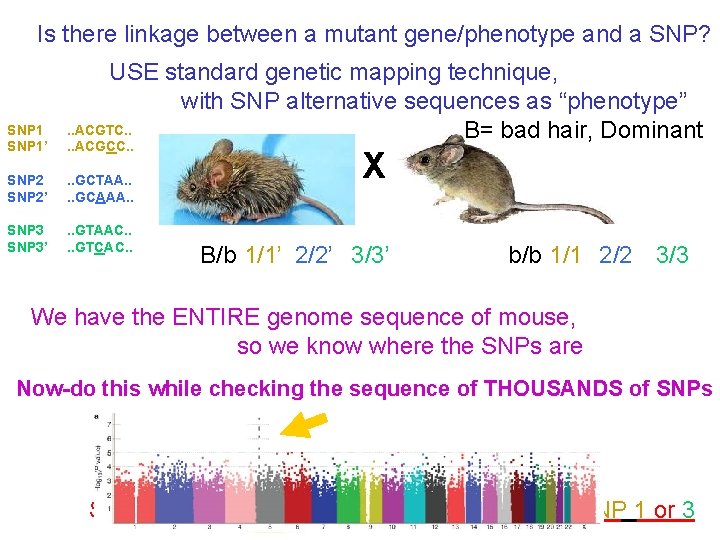
Is there linkage between a mutant gene/phenotype and a SNP? SNP 1’ USE standard genetic mapping technique, with SNP alternative sequences as “phenotype”. . ACGTC. . B= bad hair, Dominant. . ACGCC. . SNP 2’ . . GCTAA. . GCAAA. . SNP 3’ . . GTAAC. . GTCAC. . X B/b 1/1’ 2/2’ 3/3’ b/b 1/1 2/2 3/3 We have the ENTIRE genome sequence of mouse, so we know where the SNPs are Now-do this while checking the sequence of THOUSANDS of SNPs SO…B is 6 c. M from SNP 2, and is unlinked to SNP 1 or 3

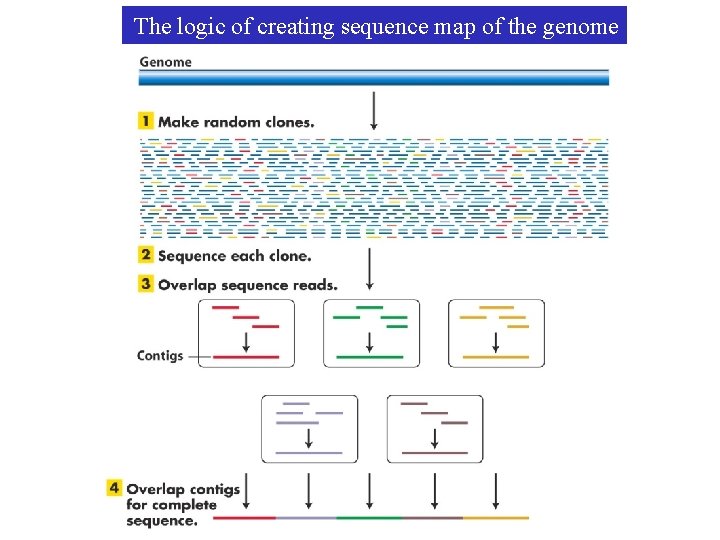
The logic of creating sequence map of the genome

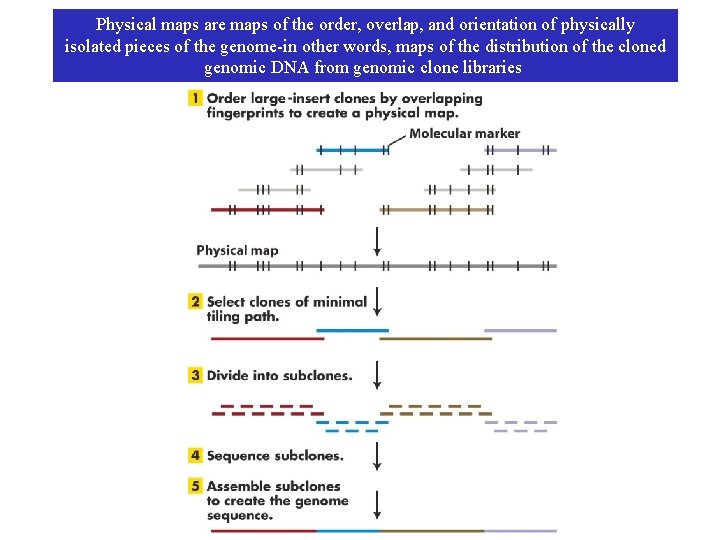
Physical maps are maps of the order, overlap, and orientation of physically isolated pieces of the genome-in other words, maps of the distribution of the cloned genomic DNA from genomic clone libraries

Part of the automated production line of a major human genome sequencing center

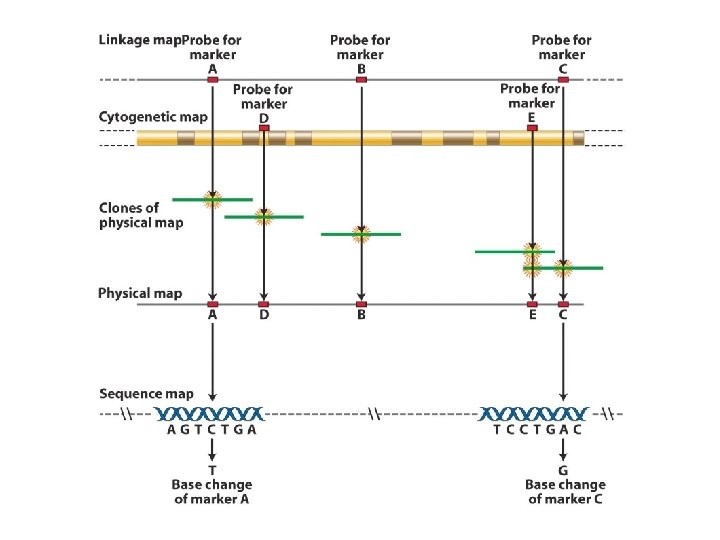
A specific gene can be found in the genomic sequence by matching linkage and cytological maps with the Genome sequence





CGCGCG vs. CGCG




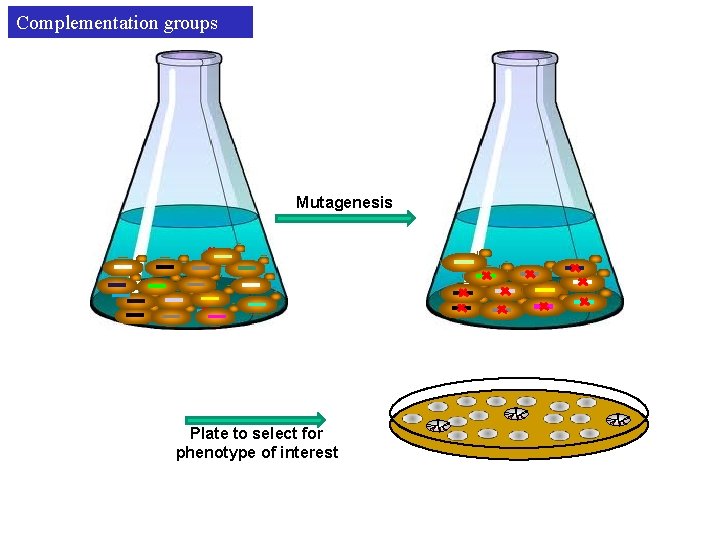
Complementation groups Mutagenesis Plate to select for phenotype of interest

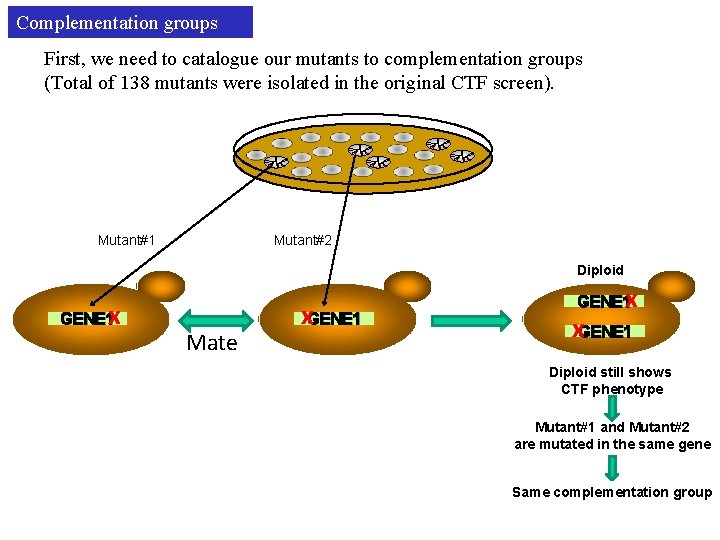
Complementation groups First, we need to catalogue our mutants to complementation groups (Total of 138 mutants were isolated in the original CTF screen). Mutant#1 Mutant#2 Diploid x GENE 1 Mate x. GENE 1 Diploid still shows CTF phenotype Mutant#1 and Mutant#2 are mutated in the same gene Same complementation group

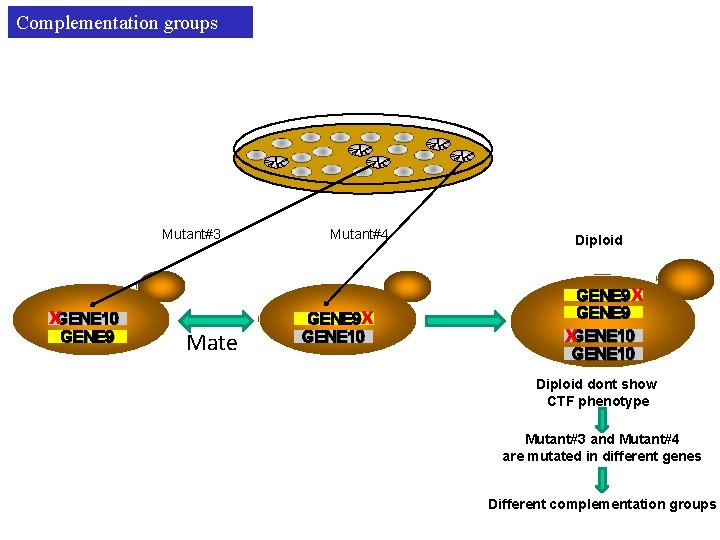
Complementation groups Mutant#3 x. GENE 10 GENE 9 Mate Mutant#4 x GENE 9 GENE 10 Diploid x GENE 9 GENE 10 x Diploid dont show CTF phenotype Mutant#3 and Mutant#4 are mutated in different genes Different complementation groups

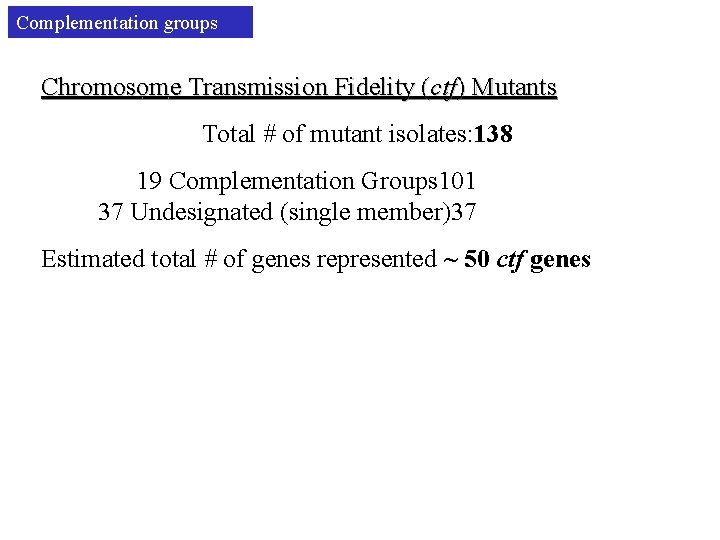
Complementation groups Chromosome Transmission Fidelity (ctf) Mutants Total # of mutant isolates: 138 19 Complementation Groups 101 37 Undesignated (single member)37 Estimated total # of genes represented ~ 50 ctf genes


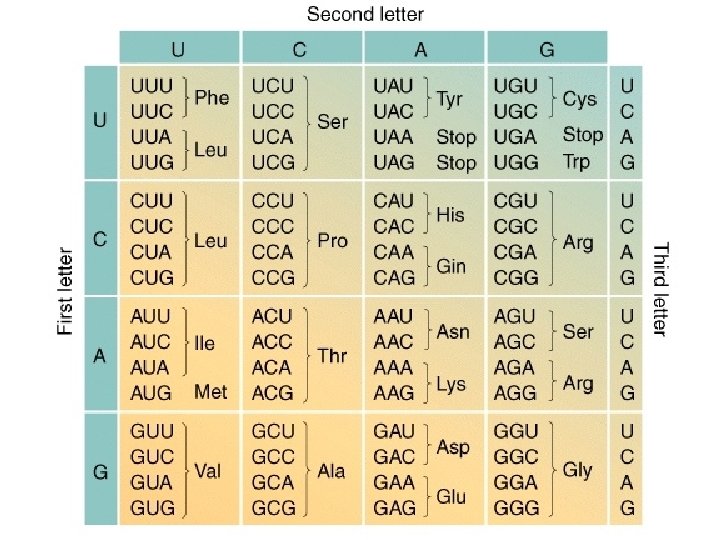
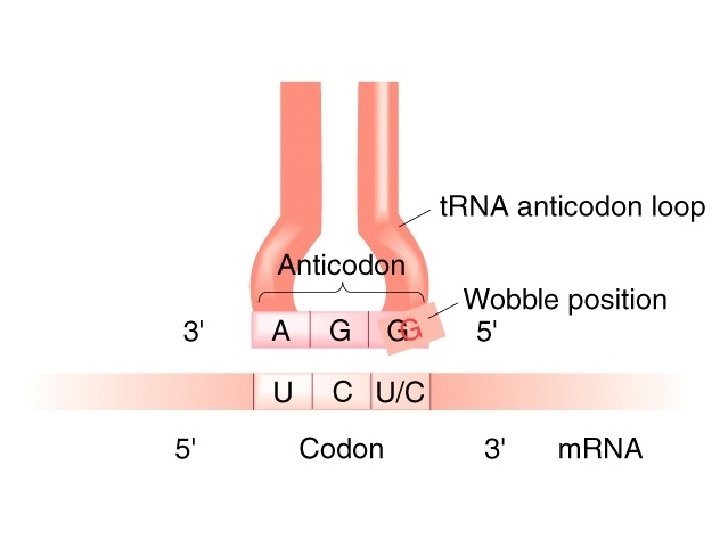
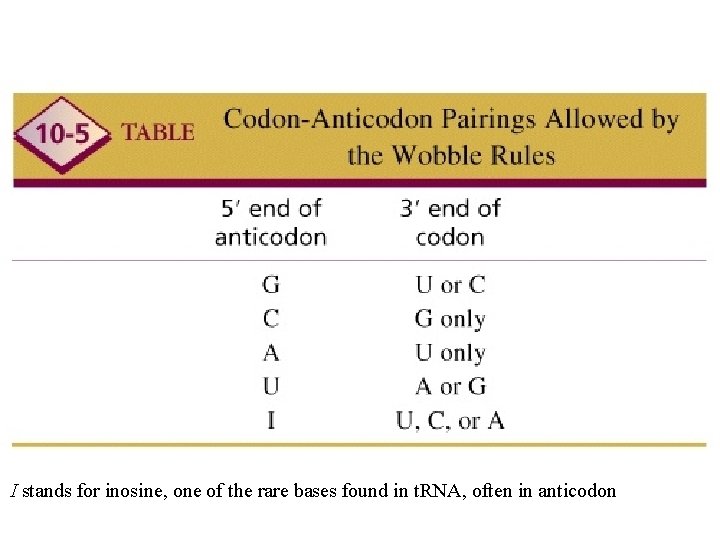
WOBBLE A situation in which the third nucleotide of an anticodon (at the 5’ end) can form two alignments. This third nucleotide can form hydrogen bonds not only with its Normal complementary nucleotide in the third position but Also with different nucleotide in the position.


I stands for inosine, one of the rare bases found in t. RNA, often in anticodon


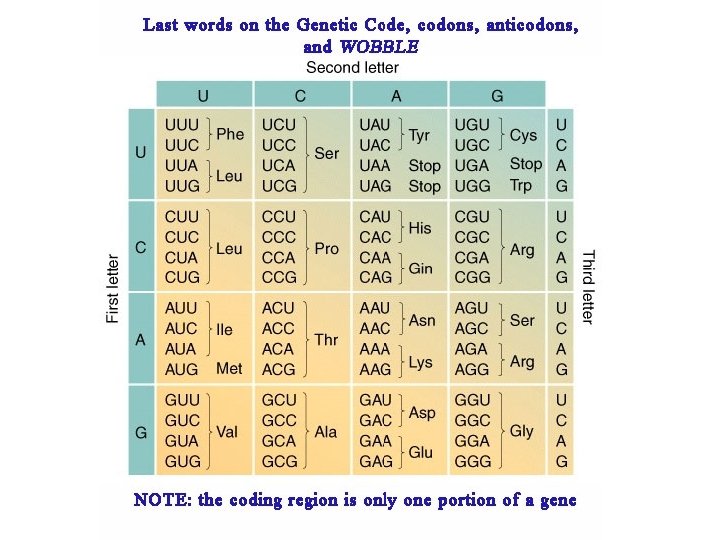
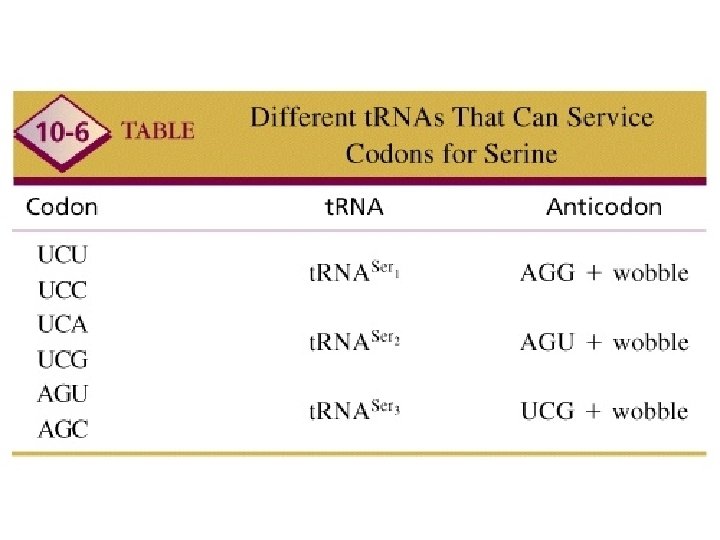
MESSAGE The genetic code is said to be degenerate because in many cases more then one codon is assigned to a single amino acid, and, in addition, several codons can pair with more then on anticodon (wobble)