8 Machine learning performance assessment measures and good





- Slides: 24

8. Machine learning – performance assessment, measures and good practices Mauno Vihinen Protein Structure and Bioinformatics group Department of Experimental Medical Science and Lund University Bioinformatics Infrastructure Lund University Sweden http: //structure. bmc. lu. se

Machine learning Artificial intelligence (AI) Variation effects are complicated and complex, thus ML methods suitable in finding generalizable properties not apparent for humans Substantial amount of data available for certain variation types

Ask these questions before you develop a (ML) prediction method What is the application area? Are there data available? Do the data capture what you are after? Are the data of high quality? Are there benchmarks to assess method performance? Are systematics available? Are there existing tools? If so, could you make it better?

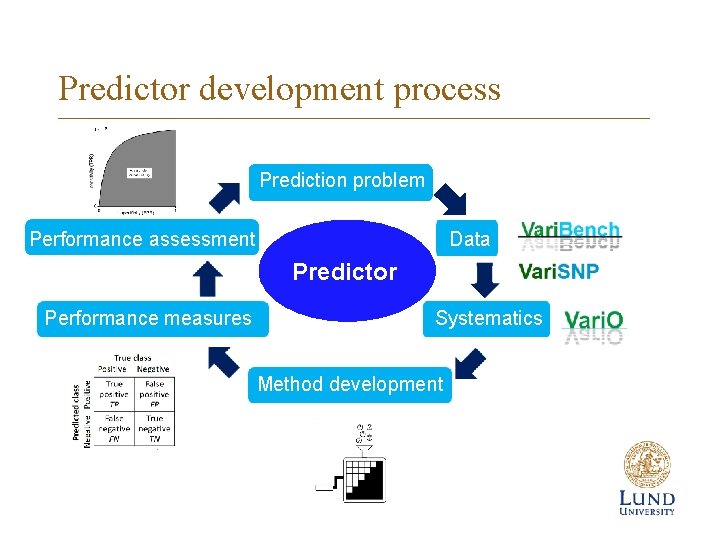
Predictor development process Prediction problem Performance assessment Data Predictor Performance measures Systematics Method development

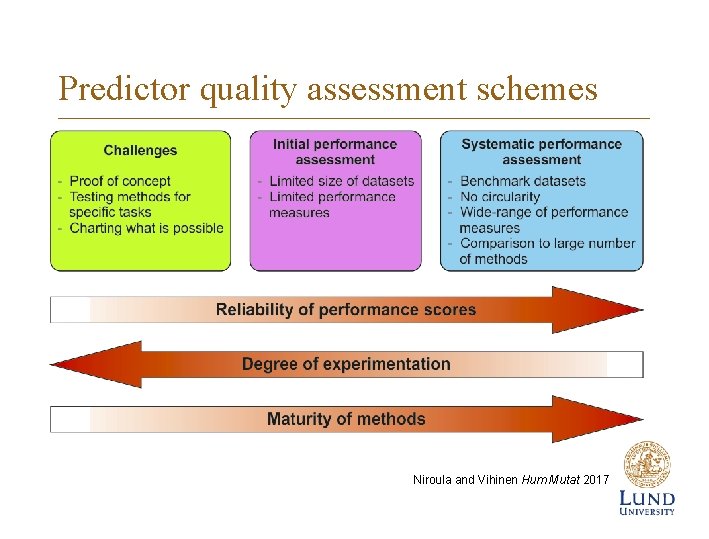
Predictor quality assessment schemes Niroula and Vihinen Hum Mutat 2017

CAGI Critical Assessment contests CASP, CAFA, CAPRI etc. Critical Assessment of Genome Interpretation, 5 challenges this far https: //genomeinterpretation. org/ Community wide challenges on different prediction tasks Blind prediction and assessment based on experimental data Special issues in Human Mutation in 2017 and 2019 summarizing results from several rounds of challenges

Variation data and benchmarks, Vari. Benchmarks for variation effects Datasets with experimentally verified outcome Used as gold standards e. g. for prediction method performance assessment and predictor development Variants are mapped to DNA/RNA/protein/3 D structure Widely adapted by the community http: //structure. bmc. lu. se/Vari. Bench Nair and Vihinen Hum. Mutat 2013

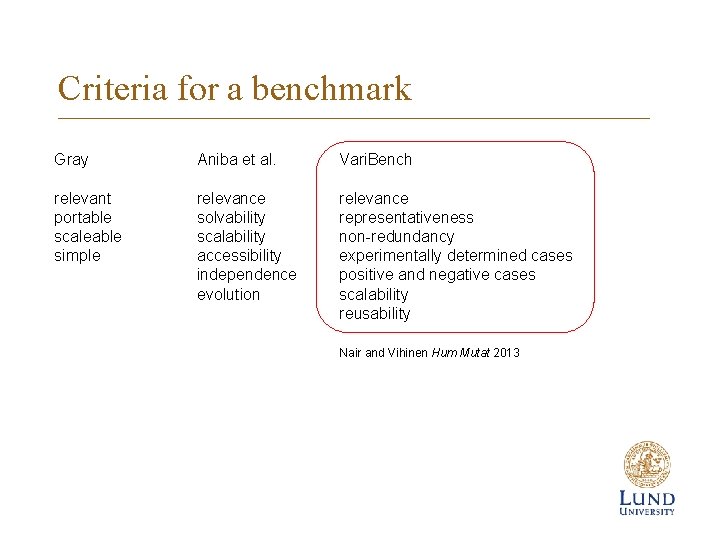
Criteria for a benchmark Gray Aniba et al. Vari. Bench relevant portable scaleable simple relevance solvability scalability accessibility independence evolution relevance representativeness non-redundancy experimentally determined cases positive and negative cases scalability reusability Nair and Vihinen Hum Mutat 2013

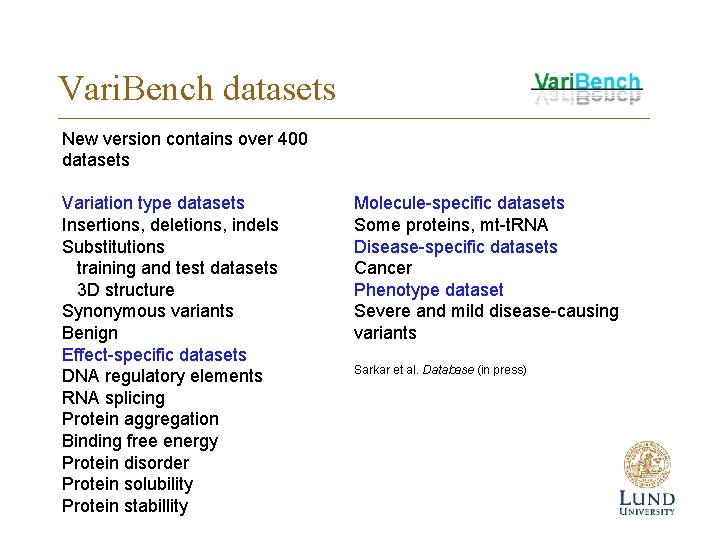
Vari. Bench datasets New version contains over 400 datasets Variation type datasets Insertions, deletions, indels Substitutions training and test datasets 3 D structure Synonymous variants Benign Effect-specific datasets DNA regulatory elements RNA splicing Protein aggregation Binding free energy Protein disorder Protein solubility Protein stabillity Molecule-specific datasets Some proteins, mt-t. RNA Disease-specific datasets Cancer Phenotype dataset Severe and mild disease-causing variants Sarkar et al. Database (in press)

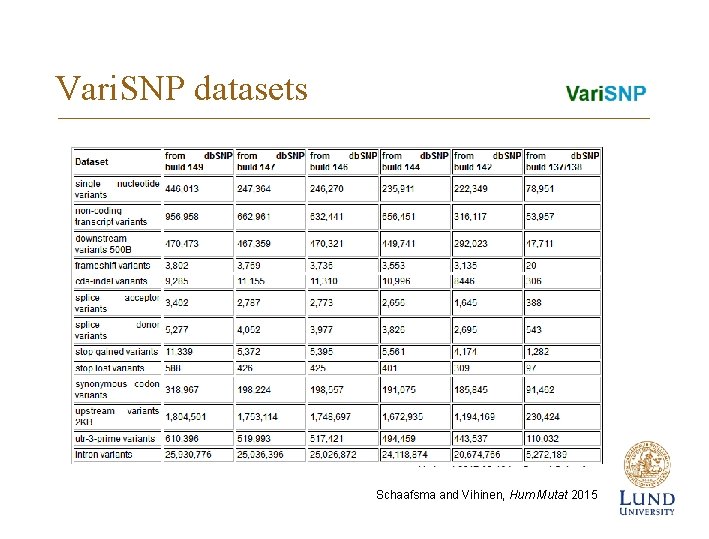
Vari. SNP datasets Schaafsma and Vihinen, Hum Mutat 2015

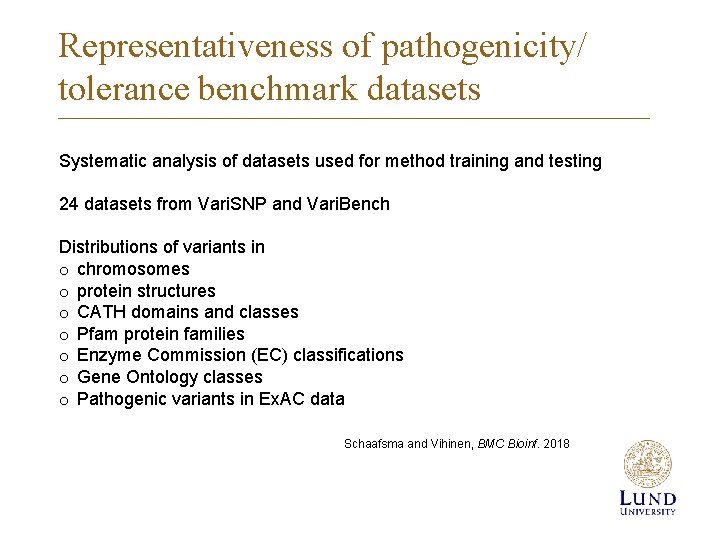
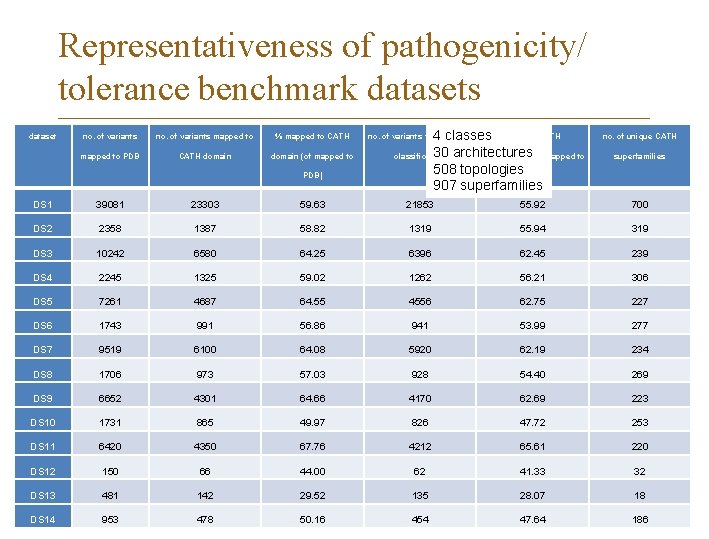
Representativeness of pathogenicity/ tolerance benchmark datasets Systematic analysis of datasets used for method training and testing 24 datasets from Vari. SNP and Vari. Bench Distributions of variants in o chromosomes o protein structures o CATH domains and classes o Pfam protein families o Enzyme Commission (EC) classifications o Gene Ontology classes o Pathogenic variants in Ex. AC data Schaafsma and Vihinen, BMC Bioinf. 2018

Representativeness of pathogenicity/ tolerance benchmark datasets dataset no. of variants mapped to % mapped to CATH mapped to PDB CATH domain (of mapped to PDB) 4 classes % with a CATH 30 architectures classification (of mapped to 508 topologies. PDB) 907 superfamilies no. of variants with a CATH no. of unique CATH superfamilies DS 1 39081 23303 59. 63 21853 55. 92 700 DS 2 2358 1387 58. 82 1319 55. 94 319 DS 3 10242 6580 64. 25 6396 62. 45 239 DS 4 2245 1325 59. 02 1262 56. 21 306 DS 5 7261 4687 64. 55 4556 62. 75 227 DS 6 1743 991 56. 86 941 53. 99 277 DS 7 9519 6100 64. 08 5920 62. 19 234 DS 8 1706 973 57. 03 928 54. 40 269 DS 9 6652 4301 64. 66 4170 62. 69 223 DS 10 1731 865 49. 97 826 47. 72 253 DS 11 6420 4350 67. 76 4212 65. 61 220 DS 12 150 66 44. 00 62 41. 33 32 DS 13 481 142 29. 52 135 28. 07 18 DS 14 953 478 50. 16 454 47. 64 186

Representativeness of pathogenicity/ tolerance benchmark datasets Schaafsma and Vihinen, BMC Bioinf. 2018

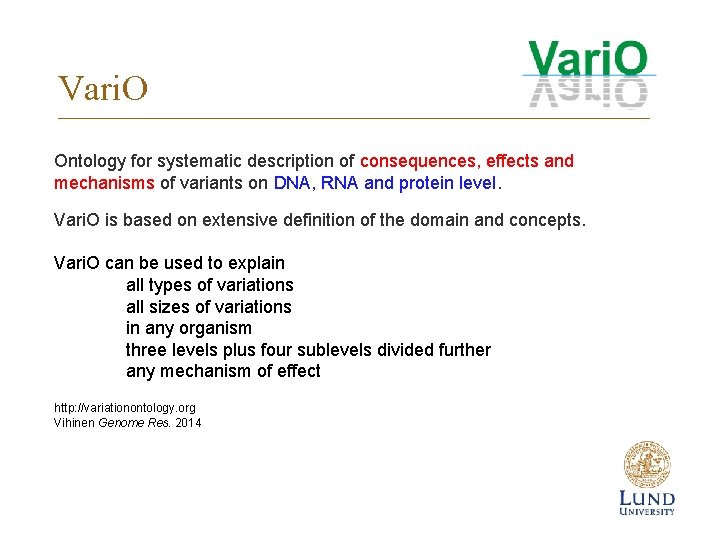
Vari. O Ontology for systematic description of consequences, effects and mechanisms of variants on DNA, RNA and protein level. Vari. O is based on extensive definition of the domain and concepts. Vari. O can be used to explain all types of variations all sizes of variations in any organism three levels plus four sublevels divided further any mechanism of effect http: //variationontology. org Vihinen Genome Res. 2014

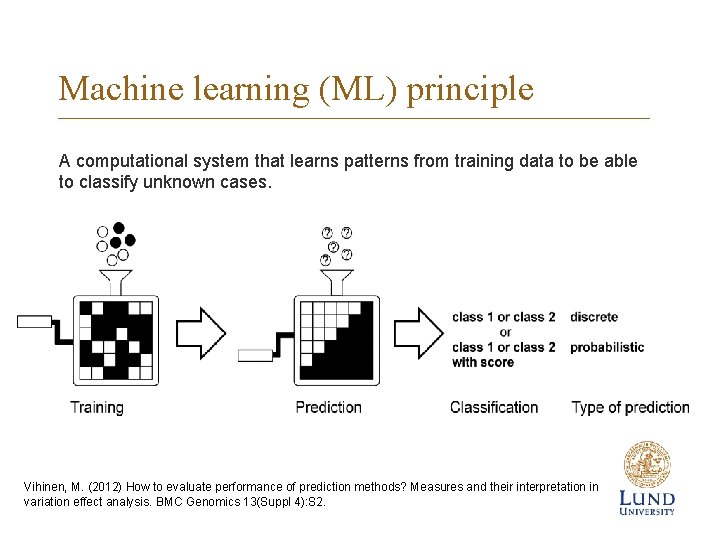
Machine learning (ML) principle A computational system that learns patterns from training data to be able to classify unknown cases. Vihinen, M. (2012) How to evaluate performance of prediction methods? Measures and their interpretation in variation effect analysis. BMC Genomics 13(Suppl 4): S 2.

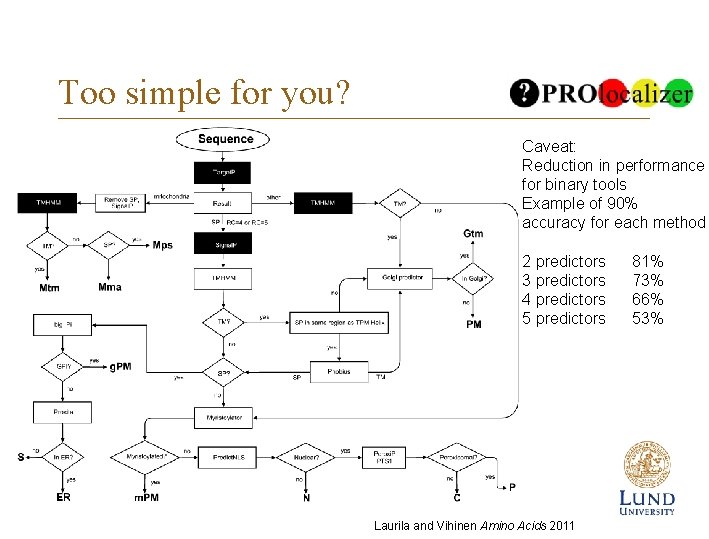
Too simple for you? Caveat: Reduction in performance for binary tools Example of 90% accuracy for each method 2 predictors 3 predictors 4 predictors 5 predictors Laurila and Vihinen Amino Acids 2011 81% 73% 66% 53%

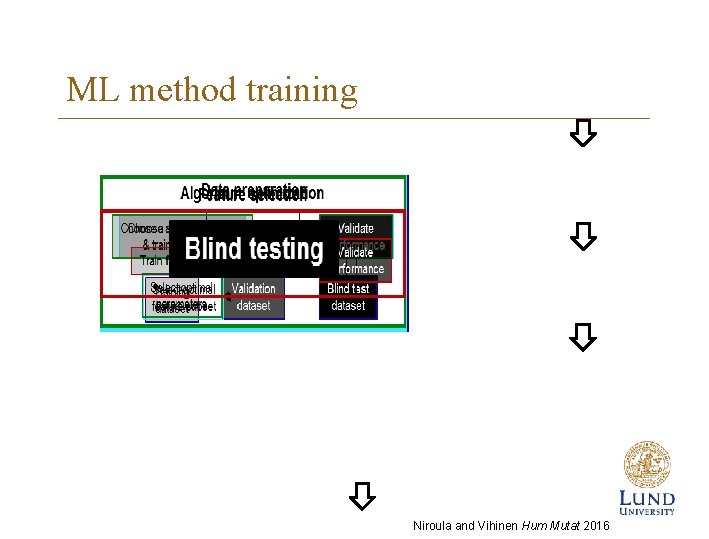
ML method training Niroula and Vihinen Hum Mutat 2016

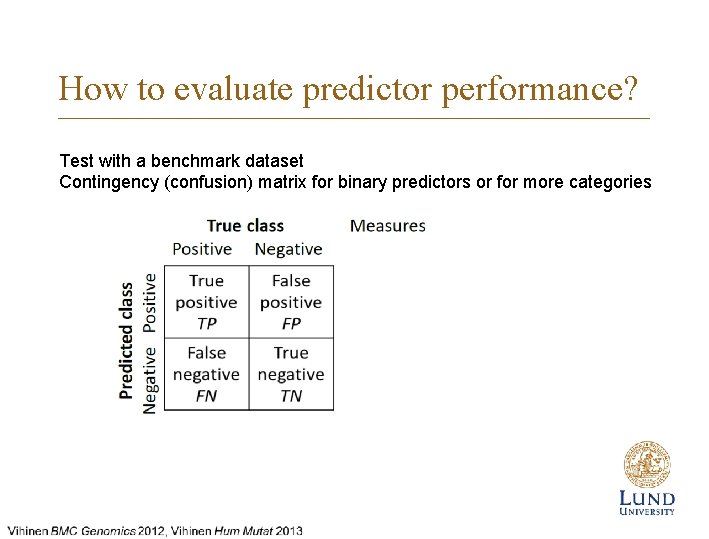
How to evaluate predictor performance? Test with a benchmark dataset Contingency (confusion) matrix for binary predictors or for more categories

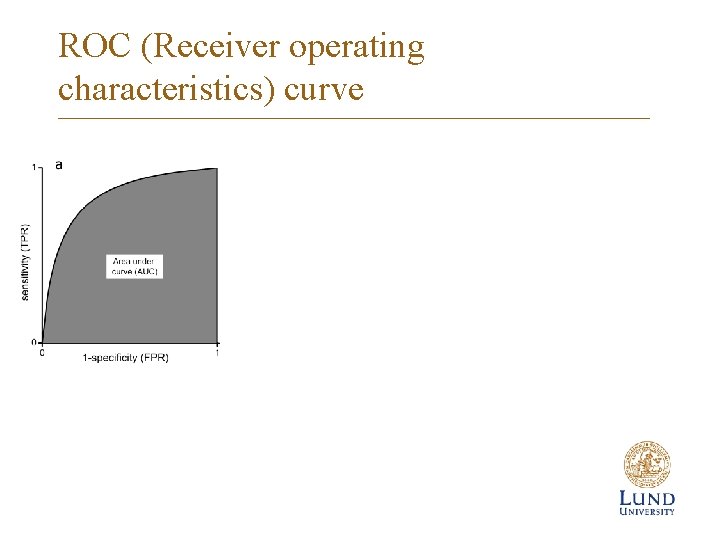
ROC (Receiver operating characteristics) curve

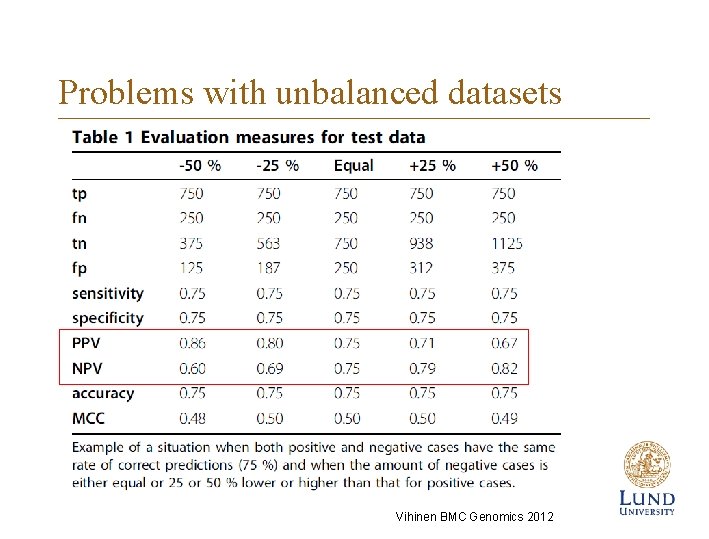
Problems with unbalanced datasets Vihinen BMC Genomics 2012

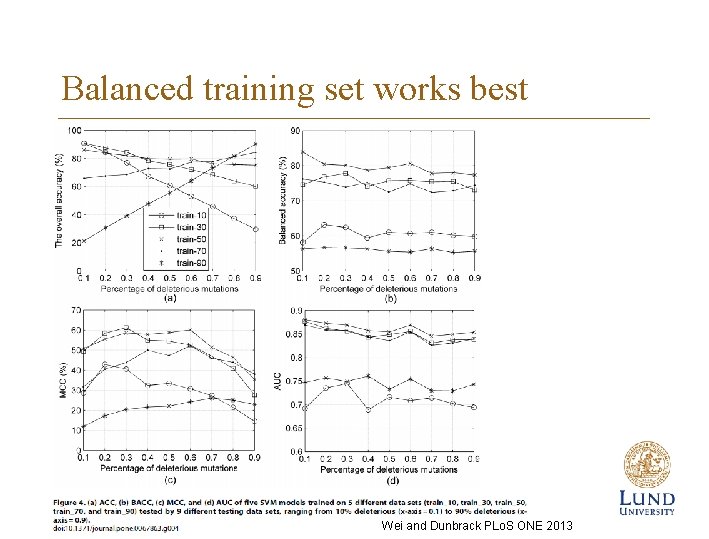
Balanced training set works best Wei and Dunbrack PLo. S ONE 2013

Measures for methods with ≥ 3 classes Several measures available, such as PCC, Pearson correlation coefficient RMSE, root mean square error MAE, mean absolute error MSE, mean squared error R 2, coefficient of determination Provide contingency table

How to report predictor and performance? Publication guidelines General requirements - the method should be novel and have reasonable performance - or have clearly improved performance compared to previous version and/or related methods - or describe novel useful parameters or applications and have reasonable performance. Method description - the method has to be described in sufficient detail - depending on the approach, the following information should be provided: description of training, optimization, input parameters and their selection, details of applied computational tools and hardware requirements - estimation of time complexity Datasets - description of training and test datasets - benchmark datasets should be used, if available - use largest available benchmark dataset - description of how data was collected, quality controls, details of sources - distribution of datasets is recommended, preferably via benchmark database - report sizes of datasets Vihinen, M. Guidelines for reporting and using prediction tools. Hum. Mutat. 2013

Predictor reporting guidelines, cont. Method performance assessment - report all appropriate performance measures - in case of binary classifier, use all six measures characterizing the contingency matrix. If possible, perform ROC analysis and provide AUC value. - perform statistically valid evaluation with sufficiently large and unbiased dataset(s) that contains both positive and negative cases. - use cross validation or some other method to separate training and test sets - training and test sets have to be disjoint - if the dataset imbalanced, make sure to use appropriate methods to mitigate this - perform comparison to related methods Implementation - detailed description of the method including user guidelines and examples (can be provided on web site) - availability of the program, either to download or as web service - batch submission possibility highly recommended - make the program preferably open source