8 1 Major Modes of Regulation Gene expression



- Slides: 39

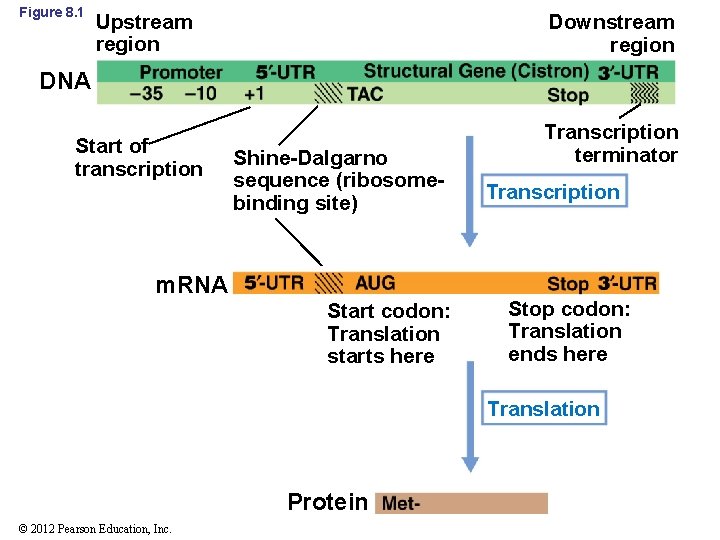
8. 1 Major Modes of Regulation • Gene expression: transcription of gene into m. RNA followed by translation of m. RNA into protein (Figure 8. 1) • Most proteins are enzymes that carry out biochemical reactions • Constitutive proteins are needed at the same level all the time • Microbial genomes encode many proteins that are not needed all the time • Regulation helps conserve energy and resources © 2012 Pearson Education, Inc.

Figure 8. 1 Upstream region Downstream region DNA Start of transcription Shine-Dalgarno sequence (ribosomebinding site) m. RNA Start codon: Translation starts here Transcription terminator Transcription Stop codon: Translation ends here Translation Protein © 2012 Pearson Education, Inc.

8. 2 DNA-Binding Proteins • m. RNA transcripts generally have a short half-life – Prevents the production of unneeded proteins • Regulation of transcription typically requires proteins that can bind to DNA • Small molecules influence the binding of regulatory proteins to DNA – Proteins actually regulate transcription © 2012 Pearson Education, Inc.

8. 2 DNA-Binding Proteins • Most DNA-binding proteins interact with DNA in a sequence-specific manner • Specificity provided by interactions between amino acid side chains and chemical groups on the bases and sugar–phosphate backbone of DNA • Major groove of DNA is the main site of protein binding • Inverted repeats frequently are binding site for regulatory proteins © 2012 Pearson Education, Inc.

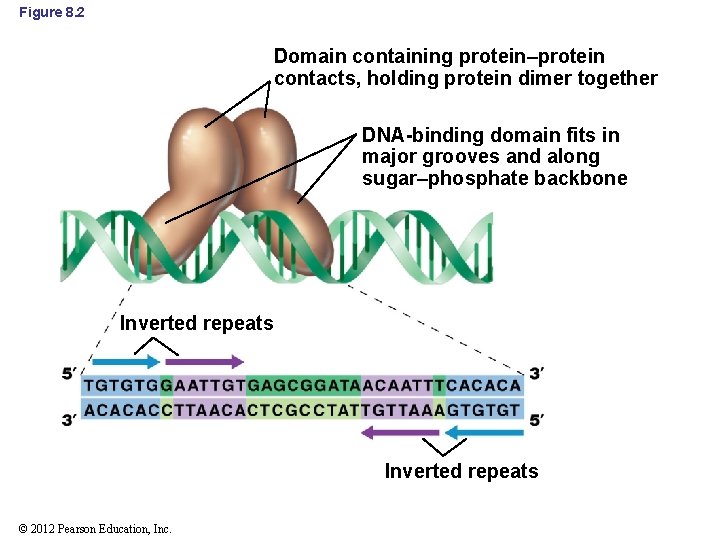
8. 2 DNA-Binding Proteins • Homodimeric proteins: proteins composed of two identical polypeptides • Protein dimers interact with inverted repeats on DNA – Each of the polypeptides binds to one inverted repeat (Figure 8. 2) © 2012 Pearson Education, Inc.

Figure 8. 2 Domain containing protein–protein contacts, holding protein dimer together DNA-binding domain fits in major grooves and along sugar–phosphate backbone Inverted repeats © 2012 Pearson Education, Inc.

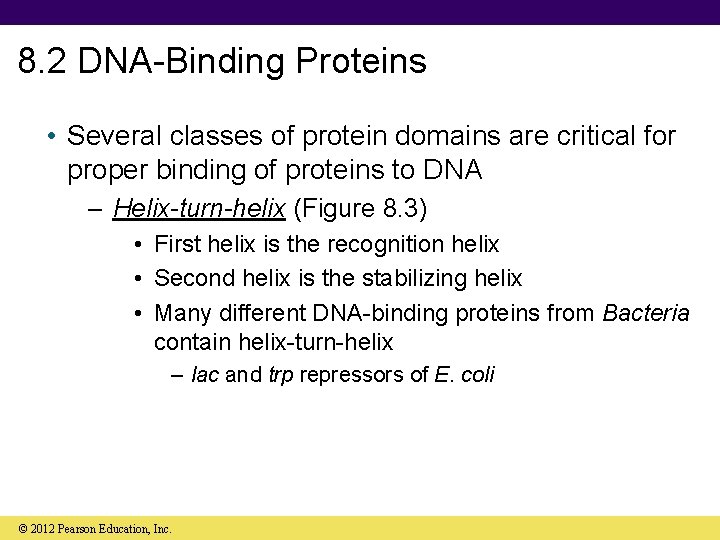
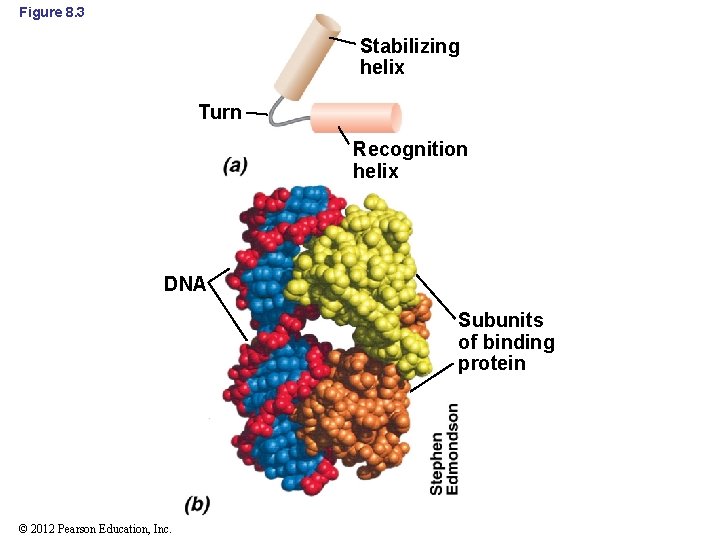
8. 2 DNA-Binding Proteins • Several classes of protein domains are critical for proper binding of proteins to DNA – Helix-turn-helix (Figure 8. 3) • First helix is the recognition helix • Second helix is the stabilizing helix • Many different DNA-binding proteins from Bacteria contain helix-turn-helix – lac and trp repressors of E. coli © 2012 Pearson Education, Inc.

Figure 8. 3 Stabilizing helix Turn Recognition helix DNA Subunits of binding protein © 2012 Pearson Education, Inc.

8. 2 DNA-Binding Proteins • Multiple outcomes after DNA binding are possible 1. DNA-binding protein may catalyze a specific reaction on the DNA molecule (i. e. , transcription by RNA polymerase) 2. The binding event can block transcription (negative regulation) 3. The binding event can activate transcription (positive regulation) © 2012 Pearson Education, Inc.

8. 3 Negative Control of Transcription: Repression and Induction • Several mechanisms for controlling gene expression in bacteria – – – These systems are greatly influenced by environment in which the organism is growing Presence or absence of specific small molecules Interactions between small molecules and DNAbinding proteins result in control of transcription or translation © 2012 Pearson Education, Inc.

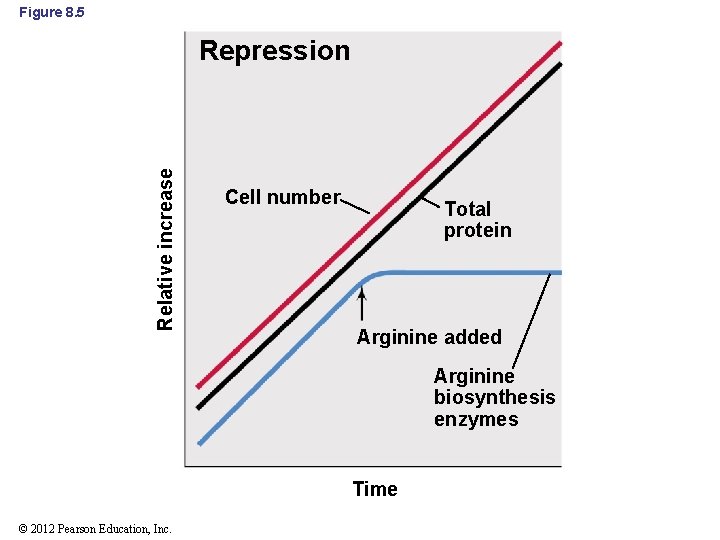
8. 3 Negative Control of Transcription: Repression and Induction • Negative control: a regulatory mechanism that stops transcription – Repression: preventing the synthesis of an enzyme in response to a signal (Figure 8. 5) • Enzymes affected by repression make up a small fraction of total proteins • Typically affects anabolic enzymes (e. g. , arginine biosynthesis) © 2012 Pearson Education, Inc.

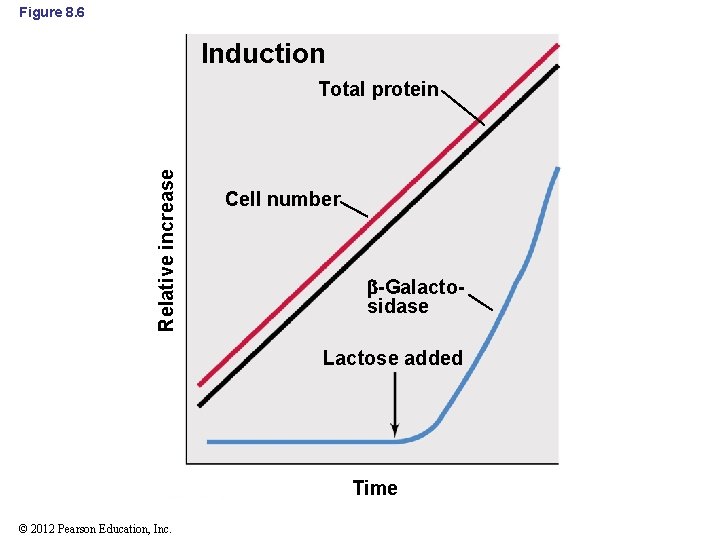
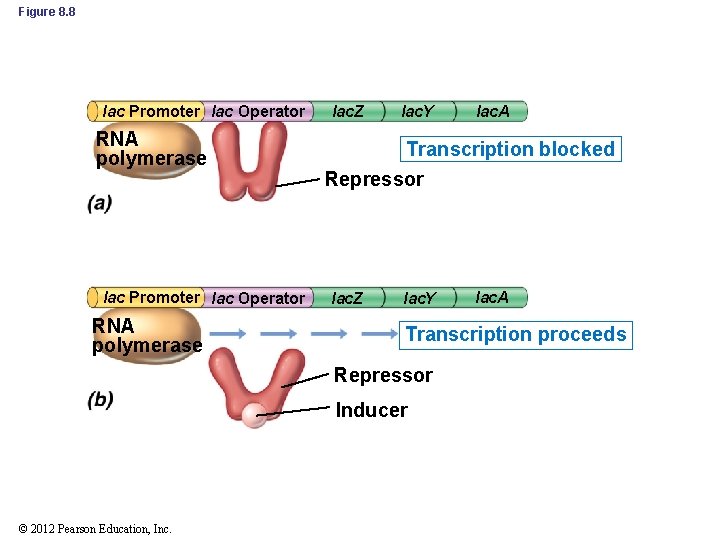
8. 3 Negative Control of Transcription: Repression and Induction • Negative Control (cont’d) – Induction: production of an enzyme in response to a signal (Figure 8. 6) • Typically affects catabolic enzymes (e. g. , lac operon) • Enzymes are synthesized only when they are needed – no wasted energy © 2012 Pearson Education, Inc.

Figure 8. 5 Relative increase Repression Cell number Total protein Arginine added Arginine biosynthesis enzymes Time © 2012 Pearson Education, Inc.

Figure 8. 6 Induction Relative increase Total protein Cell number -Galactosidase Lactose added Time © 2012 Pearson Education, Inc.

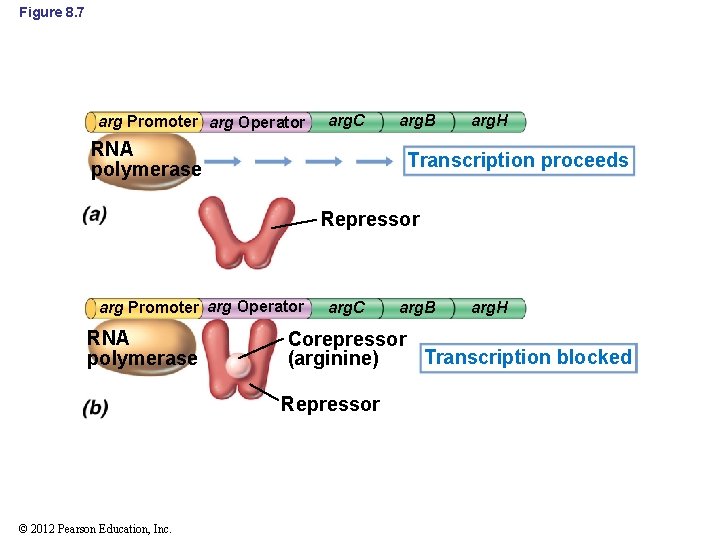
8. 3 Negative Control of Transcription: Repression and Induction • Inducer: substance that induces enzyme synthesis • Corepressor: substance that represses enzyme synthesis • Effectors: collective term for inducers and repressors • Effectors affect transcription indirectly by binding to specific DNA-binding proteins – Repressor molecules bind to an allosteric repressor protein – Allosteric repressor becomes active and binds to region of DNA near promoter called the operator © 2012 Pearson Education, Inc.

Figure 8. 7 arg Promoter arg Operator arg. C RNA polymerase arg. B arg. H Transcription proceeds Repressor arg Promoter arg Operator RNA polymerase arg. C arg. H Corepressor Transcription blocked (arginine) Repressor © 2012 Pearson Education, Inc. arg. B

Figure 8. 8 lac Promoter lac Operator RNA polymerase lac. Z lac. Y Transcription blocked Repressor lac. Z lac. Y lac. A Transcription proceeds Repressor Inducer © 2012 Pearson Education, Inc. lac. A

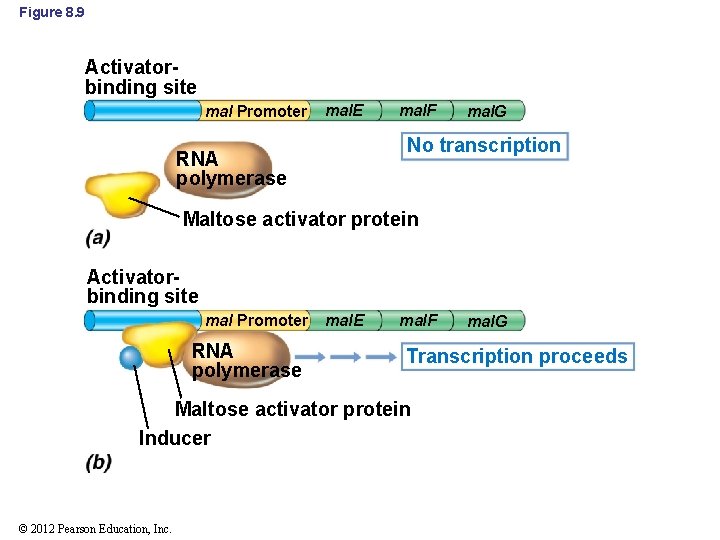
8. 4 Positive Control of Transcription • Positive control: regulator protein activates the binding of RNA polymerase to DNA (Figure 8. 9) • Maltose catabolism in E. coli – Maltose activator protein cannot bind to DNA unless it first binds maltose • Activator proteins bind specifically to certain DNA sequence – Called activator-binding site, not operator © 2012 Pearson Education, Inc.

Figure 8. 9 Activatorbinding site mal Promoter mal. E mal. F mal. G No transcription RNA polymerase Maltose activator protein Activatorbinding site mal Promoter RNA polymerase mal. E mal. F Transcription proceeds Maltose activator protein Inducer © 2012 Pearson Education, Inc. mal. G

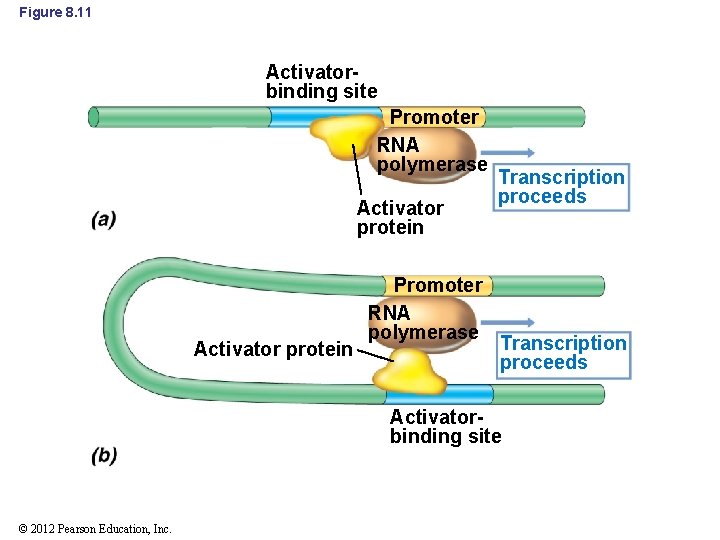
8. 4 Positive Control of Transcription • Promoters of positively controlled operons only weakly bind RNA polymerase • Activator protein helps RNA polymerase recognize promoter – May cause a change in DNA structure – May interact directly with RNA polymerase • Activator-binding site may be close to the promoter or several hundred base pairs away (Figure 8. 11) © 2012 Pearson Education, Inc.

Figure 8. 11 Activatorbinding site Promoter RNA polymerase Activator protein Promoter RNA polymerase Transcription proceeds Activatorbinding site © 2012 Pearson Education, Inc.

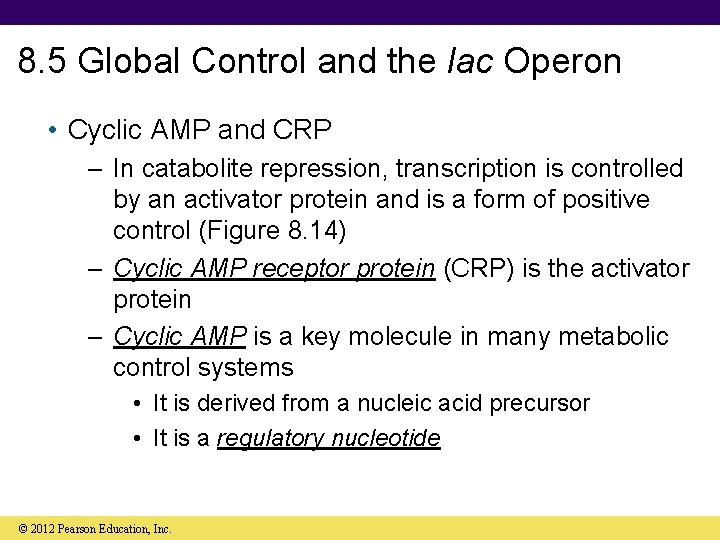
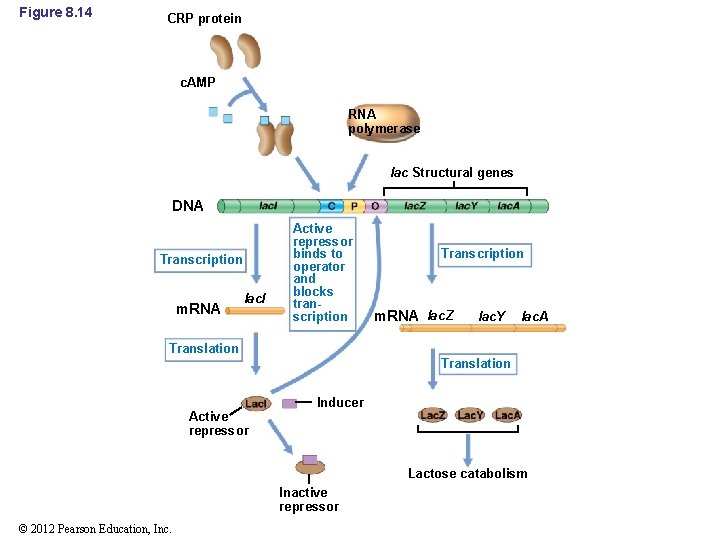
8. 5 Global Control and the lac Operon • Cyclic AMP and CRP – In catabolite repression, transcription is controlled by an activator protein and is a form of positive control (Figure 8. 14) – Cyclic AMP receptor protein (CRP) is the activator protein – Cyclic AMP is a key molecule in many metabolic control systems • It is derived from a nucleic acid precursor • It is a regulatory nucleotide © 2012 Pearson Education, Inc.

Figure 8. 14 CRP protein c. AMP RNA polymerase lac Structural genes DNA Transcription m. RNA lac. I Active repressor binds to operator and blocks transcription Transcription m. RNA lac. Z lac. Y lac. A Translation Active repressor Inducer Lactose catabolism Inactive repressor © 2012 Pearson Education, Inc.

8. 5 Global Control and the lac Operon • Dozens of catabolic operons affected by catabolite repression – Enzymes for degrading lactose, maltose, and other common carbon sources • Flagellar genes are also controlled by catabolite repression – No need to swim in search of nutrients © 2012 Pearson Education, Inc.

8. 7 Two-Component Regulatory Systems • Prokaryotes regulate cellular metabolism in response to environmental fluctuations – External signal is transmitted directly to the target – External signal detected by sensor and transmitted to regulatory machinery (Signal transduction) • Most signal transduction systems are twocomponent regulatory systems © 2012 Pearson Education, Inc.

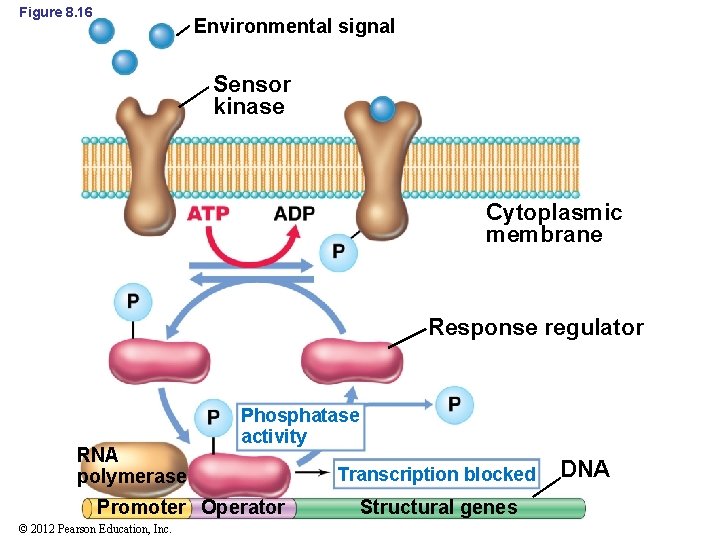
8. 7 Two-Component Regulatory Systems • Two-component regulatory systems (Figure 8. 16) – Made up of two different proteins: • Sensor kinase: (in cytoplasmic membrane) detects environmental signal and autophosphorylates • Response regulator: (in cytoplasm) DNA-binding protein that regulates transcription – Also has feedback loop • Terminates signal © 2012 Pearson Education, Inc.

Figure 8. 16 Environmental signal Sensor kinase Cytoplasmic membrane Response regulator RNA polymerase Phosphatase activity Promoter Operator © 2012 Pearson Education, Inc. Transcription blocked Structural genes DNA

8. 7 Two-Component Regulatory Systems • Almost 50 different two-component systems in E. coli – Examples include phosphate assimilation, nitrogen metabolism, and osmotic pressure response • Some signal transduction systems have multiple regulatory elements • Some Archaea also have two-component regulatory systems © 2012 Pearson Education, Inc.

8. 9 Quorum Sensing • Prokaryotes can respond to the presence of other cells of the same species • Quorum sensing: mechanism by which bacteria assess their population density – Ensures sufficient number of cells are present before initiating a response that requires a certain cell density to have an effect (e. g. , toxin production in pathogenic bacterium) © 2012 Pearson Education, Inc.

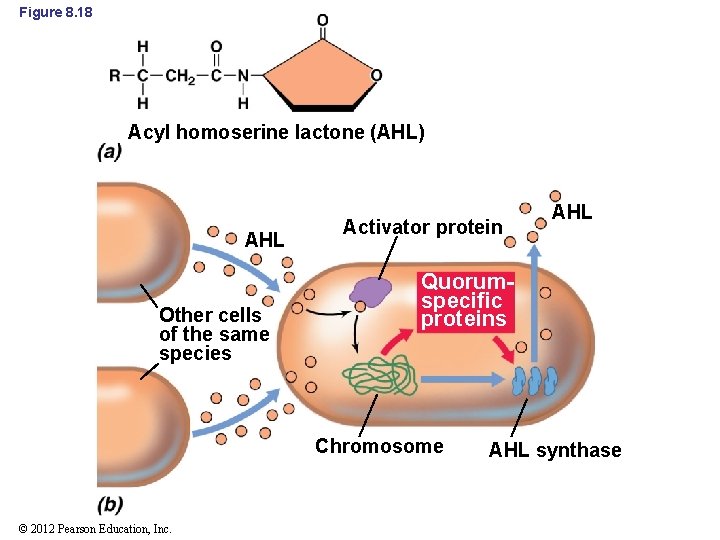
8. 9 Quorum Sensing • Each species of bacterium produces a specific autoinducer molecule (Figure 8. 18) – Diffuses freely across the cell envelope – Reaches high concentrations inside cell only if many cells are near – Binds to specific activator protein and triggers transcription of specific genes © 2012 Pearson Education, Inc.

Figure 8. 18 Acyl homoserine lactone (AHL) AHL Other cells of the same species Activator protein Quorumspecific proteins Chromosome © 2012 Pearson Education, Inc. AHL synthase

8. 9 Quorum Sensing • Several different classes of autoinducers – Acyl homoserine lactone was the first autoinducer to be identified • Quorum sensing first discovered as mechanism regulating light production in bacteria including Aliivibrio fischeri (Figure 8. 19) – Lux operon encodes bioluminescence © 2012 Pearson Education, Inc.

Figure 8. 19 © 2012 Pearson Education, Inc.

8. 9 Quorum Sensing • Examples of quorum sensing – P. aeruginosa switches from free living to growing as a biofilm – Virulence factors of Staphylococcus aureus • Quorum sensing is present in some microbial eukaryotes • Quorum sensing likely exists in Archaea © 2012 Pearson Education, Inc.

8. 11 Other Global Control Networks • Several other global control systems – Aerobic and anaerobic respiration – Catabolite repression – Nitrogen utilization – Oxidative stress – SOS response – Heat shock response © 2012 Pearson Education, Inc.

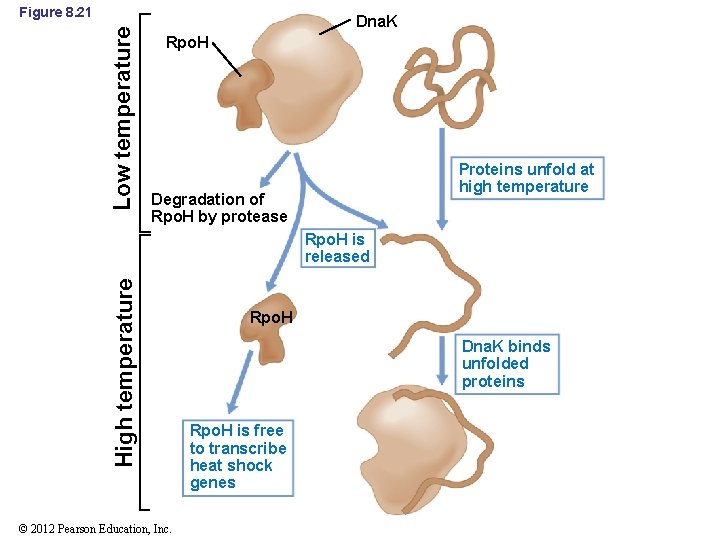
8. 11 Other Global Control Networks • Heat shock response: – Largely controlled by alternative sigma factors (Figure 8. 21) – Heat shock proteins: counteract damage of denatured proteins and help cell recover from temperature stress • Very ancient proteins • Heat shock response also occurs in Archaea © 2012 Pearson Education, Inc.

Low temperature Figure 8. 21 Dna. K Rpo. H Proteins unfold at high temperature Degradation of Rpo. H by protease High temperature Rpo. H is released © 2012 Pearson Education, Inc. Rpo. H Dna. K binds unfolded proteins Rpo. H is free to transcribe heat shock genes

IV. Regulation of Development in Model Bacteria • 8. 12 Sporulation in Bacillus • 8. 13 Caulobacter Differentiation © 2012 Pearson Education, Inc.

8. 12 Sporulation in Bacillus • Regulation of development in model bacteria – Some prokaryotes display the basic principle of differentiation • Endospore formation in Bacillus (Figure 8. 22) – Controlled by 4 sigma factors – Forms inside mother cell – Triggered by adverse external conditions (i. e. , starvation or desiccation) © 2012 Pearson Education, Inc.