7 Protein Synthesis and the Genetic Code a

- Slides: 15

7. Protein Synthesis and the Genetic Code a). Overview of translation i). Requirements for protein synthesis ii). messenger RNA iii). Ribosomes and polysomes iv). Polarity of protein synthesis b). Transfer RNA i). t. RNA as an adaptor ii). Amino acid activation iii). Aminoacyl t. RNA synthetases iv). “Charged” t. RNA c). The genetic code i). Codon-anticodon interactions ii). Initiation codon in prokaryotes vs. eukaryotes iii). Reading frame d). Mutations affecting translation i). Frameshift mutations ii). Missense and nonsense mutations

Overview of translation • last step in the flow of genetic information • definition of translation • requirements for protein synthesis • m. RNA • ribosomes • initiation factors • elongation and termination factors • GTP • aminoacyl t. RNAs • amino acids • aminoacyl t. RNA synthetases • ATP

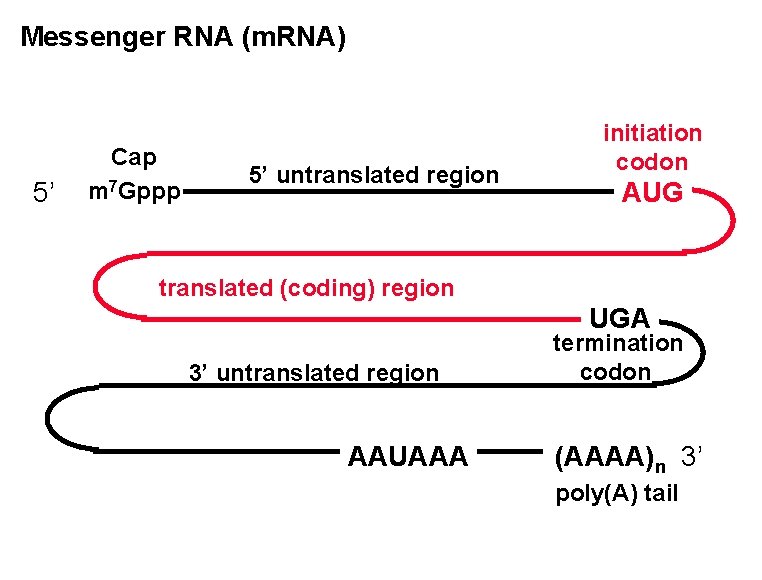
Messenger RNA (m. RNA) 5’ Cap m 7 Gppp 5’ untranslated region initiation codon AUG translated (coding) region UGA 3’ untranslated region AAUAAA termination codon (AAAA)n 3’ poly(A) tail

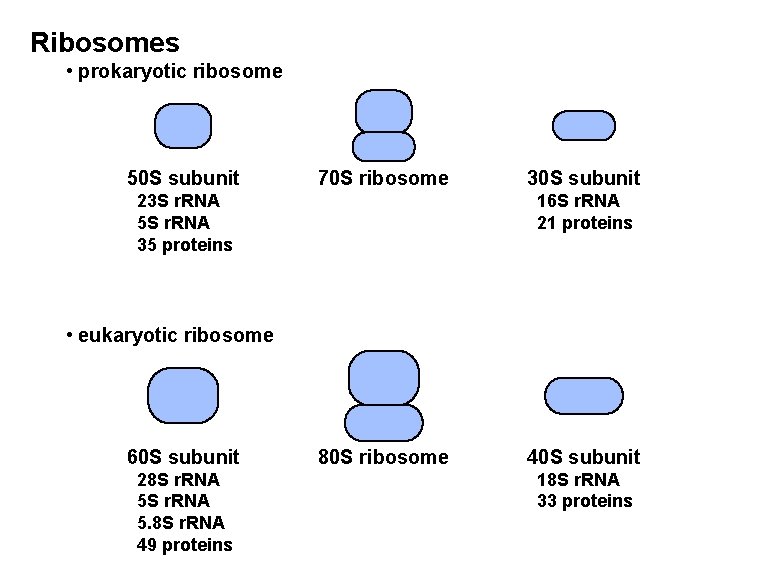
Ribosomes • prokaryotic ribosome 50 S subunit 70 S ribosome 23 S r. RNA 5 S r. RNA 35 proteins 30 S subunit 16 S r. RNA 21 proteins • eukaryotic ribosome 60 S subunit 28 S r. RNA 5. 8 S r. RNA 49 proteins 80 S ribosome 40 S subunit 18 S r. RNA 33 proteins

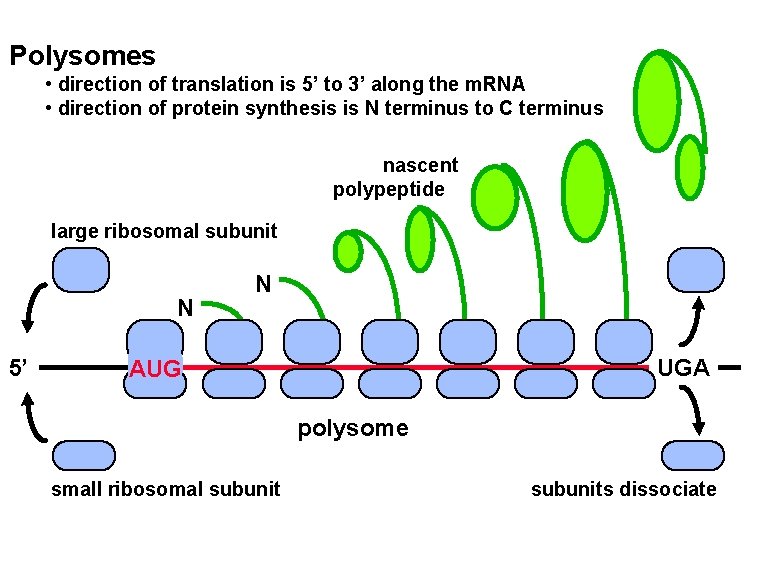
Polysomes • direction of translation is 5’ to 3’ along the m. RNA • direction of protein synthesis is N terminus to C terminus nascent polypeptide large ribosomal subunit N 5’ N UGA AUG polysome small ribosomal subunits dissociate

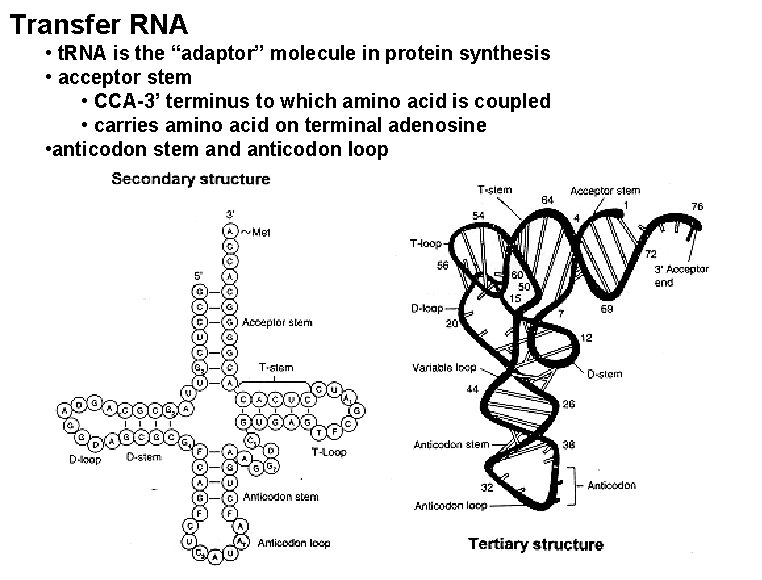
Transfer RNA • t. RNA is the “adaptor” molecule in protein synthesis • acceptor stem • CCA-3’ terminus to which amino acid is coupled • carries amino acid on terminal adenosine • anticodon stem and anticodon loop

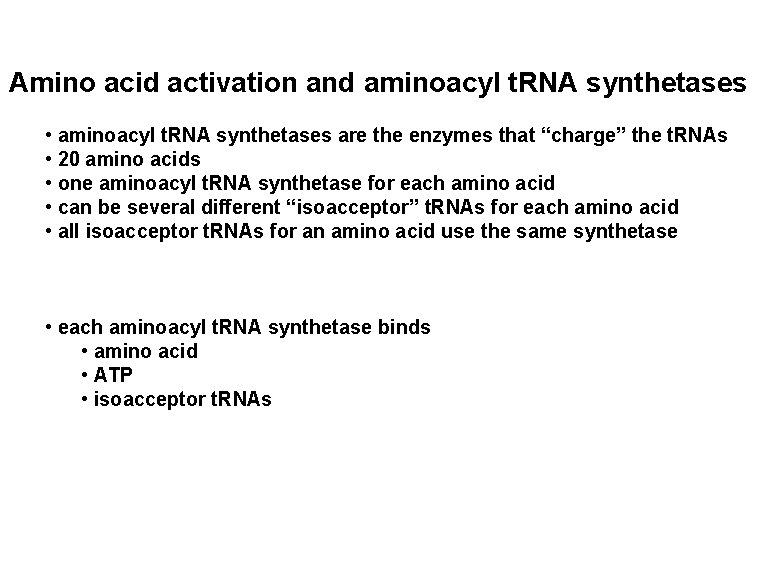
Amino acid activation and aminoacyl t. RNA synthetases • aminoacyl t. RNA synthetases are the enzymes that “charge” the t. RNAs • 20 amino acids • one aminoacyl t. RNA synthetase for each amino acid • can be several different “isoacceptor” t. RNAs for each amino acid • all isoacceptor t. RNAs for an amino acid use the same synthetase • each aminoacyl t. RNA synthetase binds • amino acid • ATP • isoacceptor t. RNAs

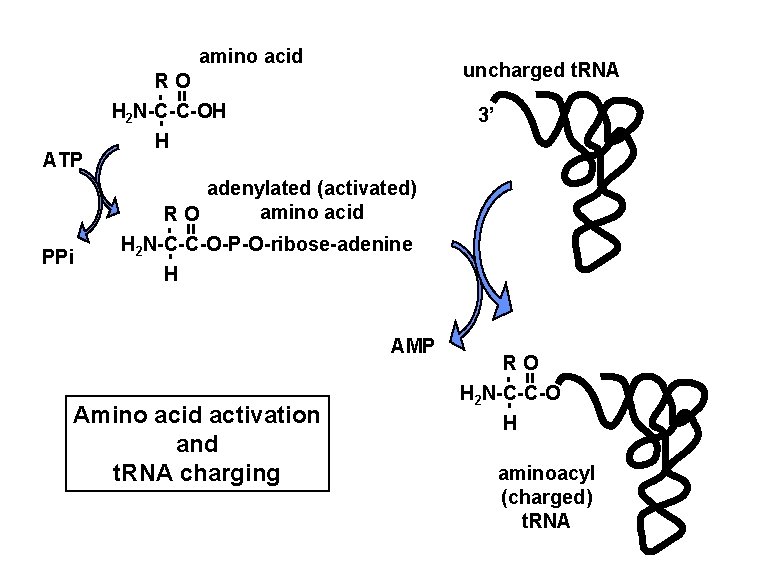
amino acid uncharged t. RNA - = RO H 2 N-C-C-OH adenylated (activated) amino acid RO H 2 N-C-C-O-P-O-ribose-adenine H AMP Amino acid activation and t. RNA charging RO H 2 N-C-C-O - = PPi H - = ATP 3’ H aminoacyl (charged) t. RNA

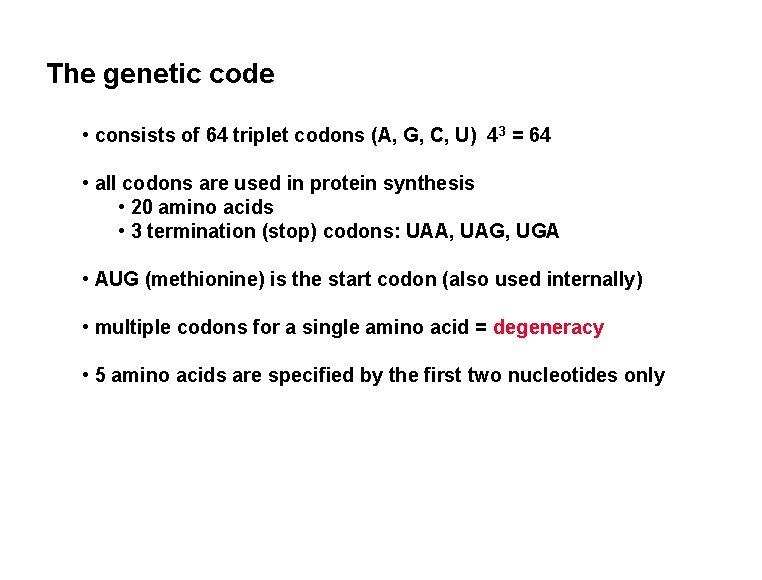
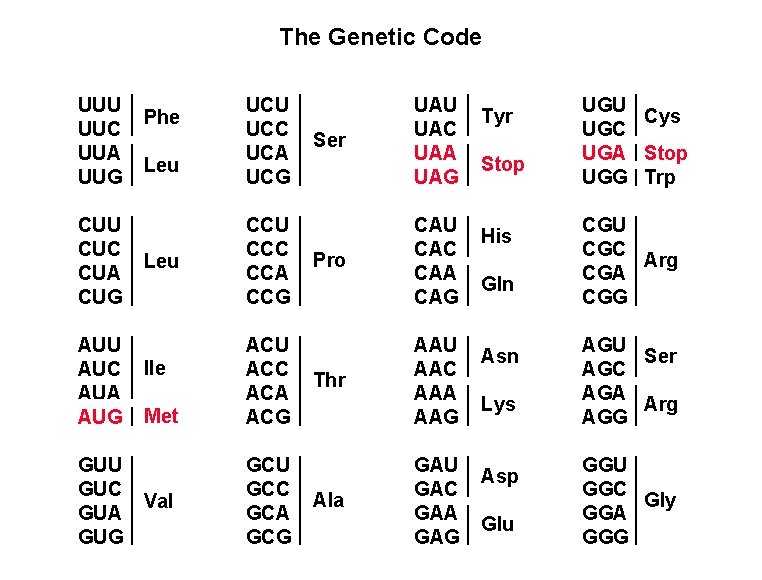
The genetic code • consists of 64 triplet codons (A, G, C, U) 43 = 64 • all codons are used in protein synthesis • 20 amino acids • 3 termination (stop) codons: UAA, UAG, UGA • AUG (methionine) is the start codon (also used internally) • multiple codons for a single amino acid = degeneracy • 5 amino acids are specified by the first two nucleotides only

The Genetic Code UUU UUC UUA UUG CUU CUC CUA CUG AUU AUC AUA AUG GUU GUC GUA GUG Phe Leu Ile Met Val UCU UCC UCA UCG CCU CCC CCA CCG ACU ACC ACA ACG GCU GCC GCA GCG Ser Pro Thr Ala UAU UAC UAA UAG CAU CAC CAA CAG AAU AAC AAA AAG GAU GAC GAA GAG Tyr Stop His Gln Asn Lys Asp Glu UGU Cys UGC UGA Stop UGG Trp CGU CGC Arg CGA CGG AGU Ser AGC AGA Arg AGG GGU GGC Gly GGA GGG

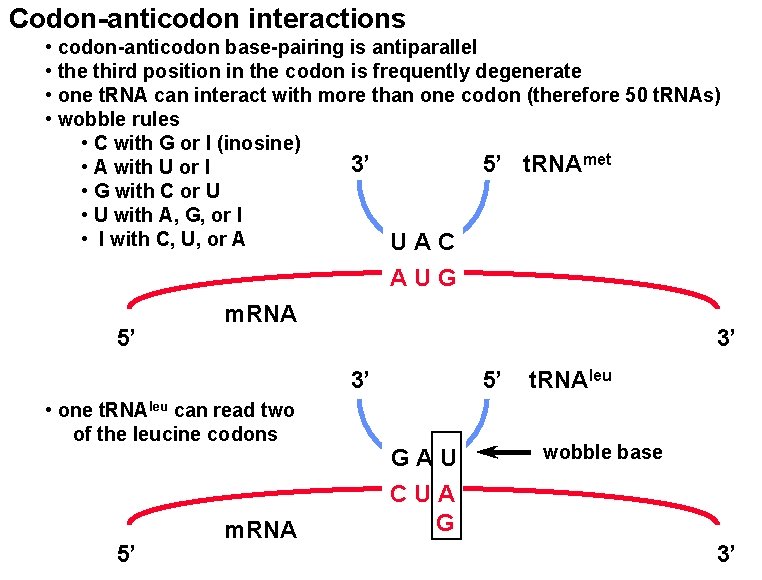
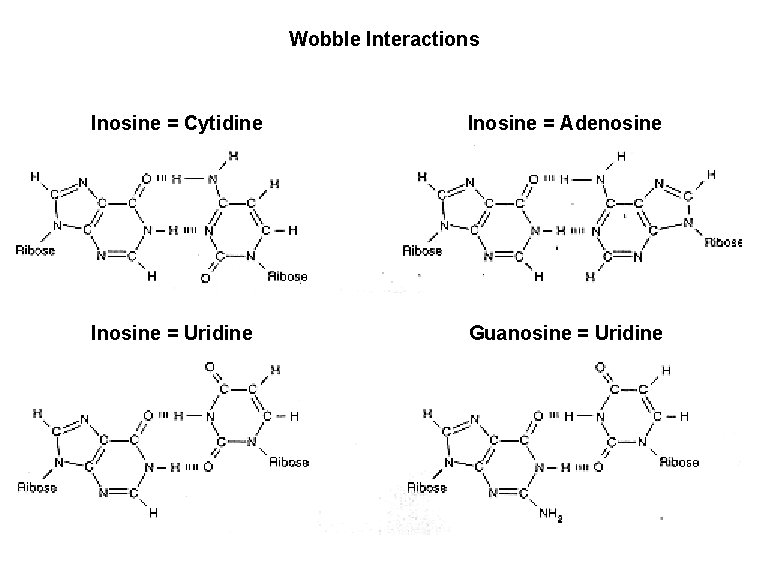
Codon-anticodon interactions • codon-anticodon base-pairing is antiparallel • the third position in the codon is frequently degenerate • one t. RNA can interact with more than one codon (therefore 50 t. RNAs) • wobble rules • C with G or I (inosine) 3’ 5’ t. RNAmet • A with U or I • G with C or U • U with A, G, or I • I with C, U, or A UAC AUG 5’ m. RNA 3’ 3’ • one t. RNAleu can read two of the leucine codons 5’ m. RNA 5’ GAU CUA G t. RNAleu wobble base 3’

Wobble Interactions Inosine = Cytidine Inosine = Adenosine Inosine = Uridine Guanosine = Uridine

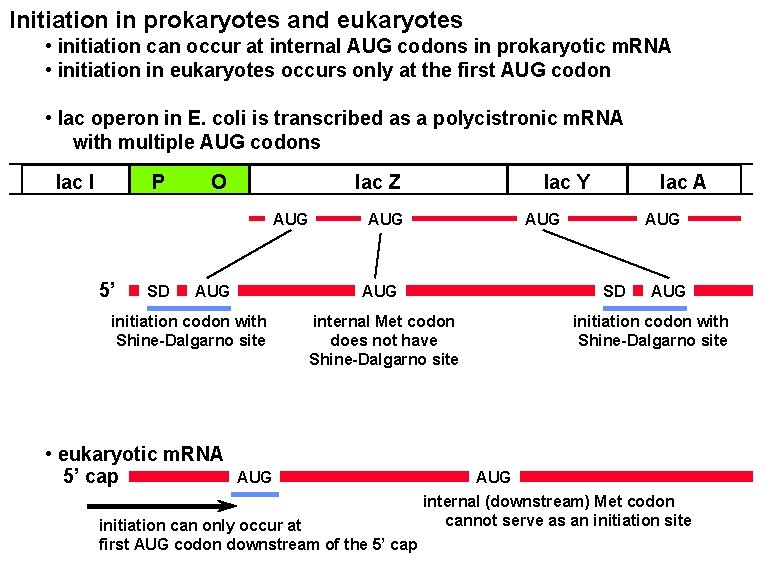
Initiation in prokaryotes and eukaryotes • initiation can occur at internal AUG codons in prokaryotic m. RNA • initiation in eukaryotes occurs only at the first AUG codon • lac operon in E. coli is transcribed as a polycistronic m. RNA with multiple AUG codons lac I P O lac Z AUG 5’ SD AUG initiation can only occur at first AUG codon downstream of the 5’ cap AUG SD internal Met codon does not have Shine-Dalgarno site AUG lac A AUG initiation codon with Shine-Dalgarno site • eukaryotic m. RNA 5’ cap lac Y AUG initiation codon with Shine-Dalgarno site AUG internal (downstream) Met codon cannot serve as an initiation site

Reading frame • reading frame is determined by the AUG initiation codon • every subsequent triplet is read as a codon until reaching a stop codon . . . AGAGCGGA. AUG. GCA. GAG. UGG. CUA. AGC. AUG. UCG. UGA. UCGAAUAAA. . . MET. ALA. GLU. TRP. LEU. SER. MET. SER • a frameshift mutation. . . AGAGCGGA. AUG. GCA. GA. UGG. CUA. AGC. AUG. UCG. UGA. UCGAAUAAA. . . • the new reading frame results in the wrong amino acid sequence and the formation of a truncated protein. . . AGAGCGGA. AUG. GCA. GAU. GGC. UAA. GCAUGUCGUGAUCGAAUAAA. . . MET. ALA. ASP. GLY

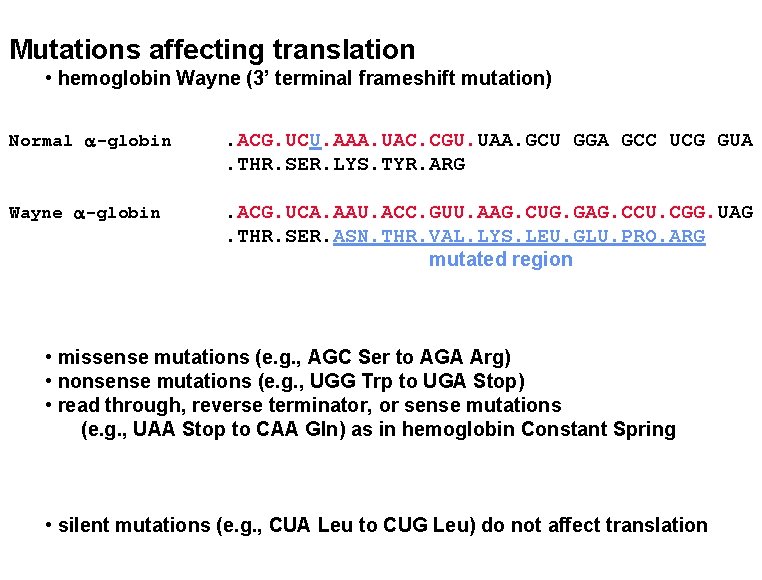
Mutations affecting translation • hemoglobin Wayne (3’ terminal frameshift mutation) Normal a-globin . ACG. UCU. AAA. UAC. CGU. UAA. GCU GGA GCC UCG GUA. THR. SER. LYS. TYR. ARG Wayne a-globin . ACG. UCA. AAU. ACC. GUU. AAG. CUG. GAG. CCU. CGG. UAG. THR. SER. ASN. THR. VAL. LYS. LEU. GLU. PRO. ARG mutated region • missense mutations (e. g. , AGC Ser to AGA Arg) • nonsense mutations (e. g. , UGG Trp to UGA Stop) • read through, reverse terminator, or sense mutations (e. g. , UAA Stop to CAA Gln) as in hemoglobin Constant Spring • silent mutations (e. g. , CUA Leu to CUG Leu) do not affect translation