7 Molecular Docking and Drug Discovery R M








- Slides: 33

7. Molecular Docking and Drug Discovery

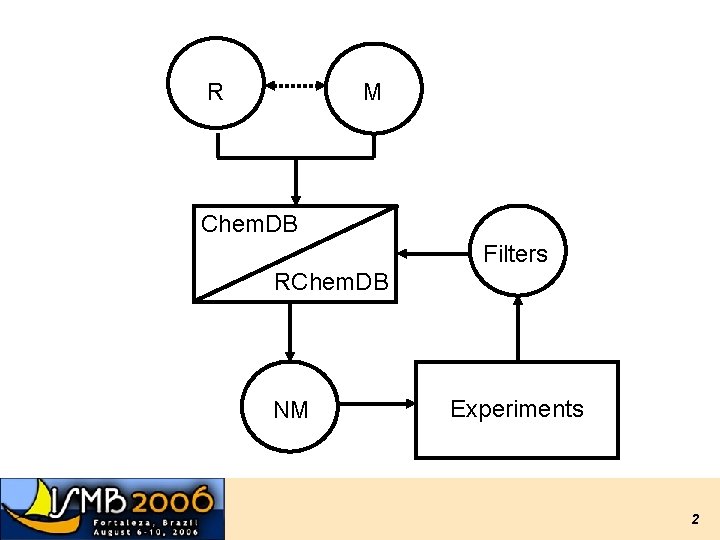
R M Chem. DB Filters RChem. DB NM Experiments 2

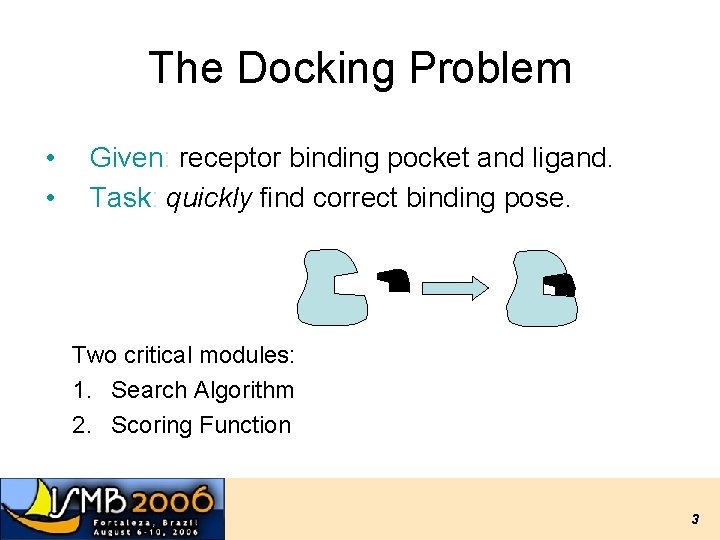
The Docking Problem • • Given: receptor binding pocket and ligand. Task: quickly find correct binding pose. Two critical modules: 1. Search Algorithm 2. Scoring Function 3

Definitions • p. Kd = measures tightness of binding • p. Ki = measures ability to inhibit • Mechanisms of action—for instance: – Competitive inhibition (most typical docking case) – Allosteric inhibition (bind to different pocket) – Allosteric activation 4

Challenges • Search algorithm – Speed (5 M compounds or more) – Local minima – High-dimensional search space • Scoring function – – – Strict control of false positives Good correlation with p. Kd Multiple terms No consensus Non-additive effects (solvation, hydrophobic interactions) • Note: p. Kd does not always correspond with activity • ADME concerns 5

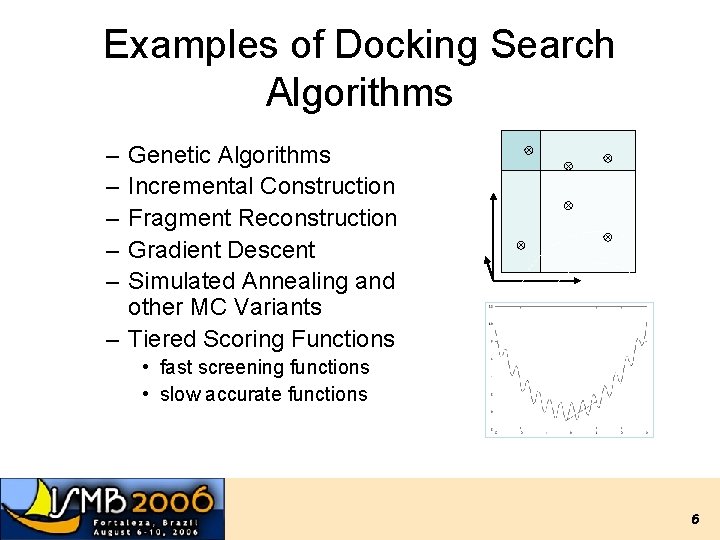
Examples of Docking Search Algorithms – – – Genetic Algorithms Incremental Construction Fragment Reconstruction Gradient Descent Simulated Annealing and other MC Variants – Tiered Scoring Functions • fast screening functions • slow accurate functions 6

High Dimensionality: Flexibility • Most algorithms handle ligand flexibility but do NOT handle receptor flexibility. • Iterative Docking to find alternate conformations of the protein – Dock flexible ligand – Minimize receptor holding ligand rigid – Repeat 7

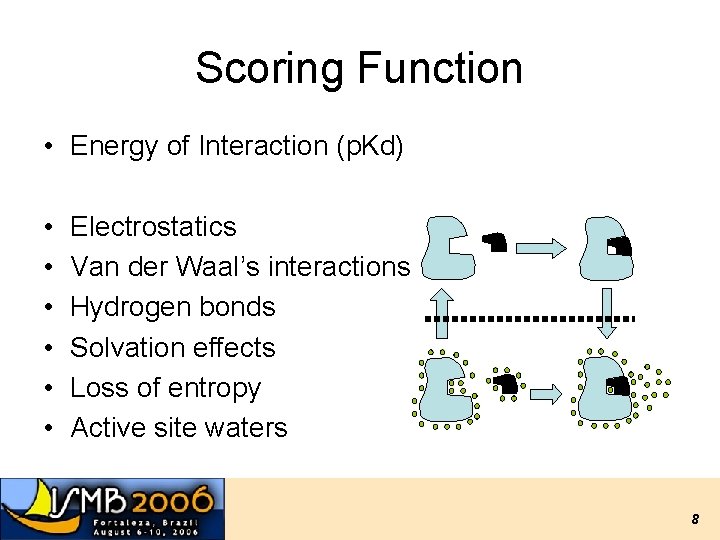
Scoring Function • Energy of Interaction (p. Kd) • • • Electrostatics Van der Waal’s interactions Hydrogen bonds Solvation effects Loss of entropy Active site waters 8

ADME concerns can be more important than bioactivity. Most of these properties are difficult to predict. • Absorption • Distribution • Metabolism • Excretion 9

Docking Programs • Dock (UCSF) • Autodock (Scripps) • Glide (Schrodinger) • ICM (Molsoft) • FRED (Open Eye) • Gold, Flex. X, etc. 10

Evaluation of Docking Programs • Evaluation of library ranking efficacy in virtual screening. J Comput Chem. 2005 Jan 15; 26(1): 11 -22. • Evaluation of docking performance: comparative data on docking algorithms. J Med Chem. 2004 Jan 29; 47(3): 558 -65. • Impact of scoring functions on enrichment in dockingbased virtual screening: an application study on renin inhibitors. J Chem Inf Comput Sci. 2004 May. Jun; 44(3): 1123 -9. 11

Cluster Based Computing • Trivially parallelizable – Divide ligand input files – Some programs have specific parallel implementations (PVM or MPI implementations, …) • Commercial licenses are expensive 12

Consensus Scoring • Combining independent scoring functions and docking algorithms can improve results • Most common method: sort using the sum of the ranks of component scores • More sophisticated methods exist Consensus scoring criteria for improving enrichment in virtual screening. J Chem Inf Model. 2005 Jul-Aug; 45(4): 1134 -46. 13

Adding Chemical Informatics Docking results can be improved by using chemical information about the hits. Chemicals which bind the same protein tend to have similar structure. Iterating back and forth between docking and searching large DB. Use other filters and predictive modules (e. g. Lipinski rules) ALGORITHM: 1. Dock and rank a chemical database 2. Create a bayesian model of the fingerprints of the top hits. 3. Re-rank the databased on their likelihood according to the bayesian model • Finding More Needles in the Haystack: A Simple and Efficient Method for Improving High. Throughput Docking Results J. Med. Chem. , 47 (11), 2743 -2749, 2004. 14

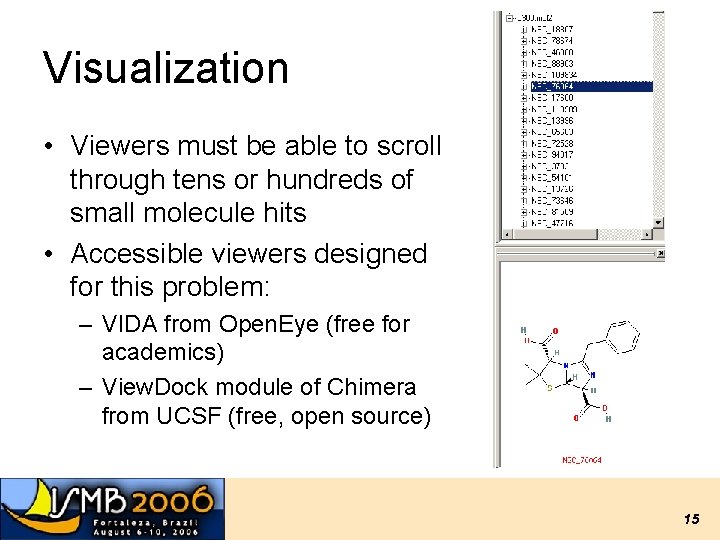
Visualization • Viewers must be able to scroll through tens or hundreds of small molecule hits • Accessible viewers designed for this problem: – VIDA from Open. Eye (free for academics) – View. Dock module of Chimera from UCSF (free, open source) 15

Long-term Goal of Drug Discovery • LTDD (Low Throughput Drug Design) instead of HTVS (High Throughput Virtual Screening) • Common ground: explore virtual space 16

Drug Discovery Case Study: Tuberculosis

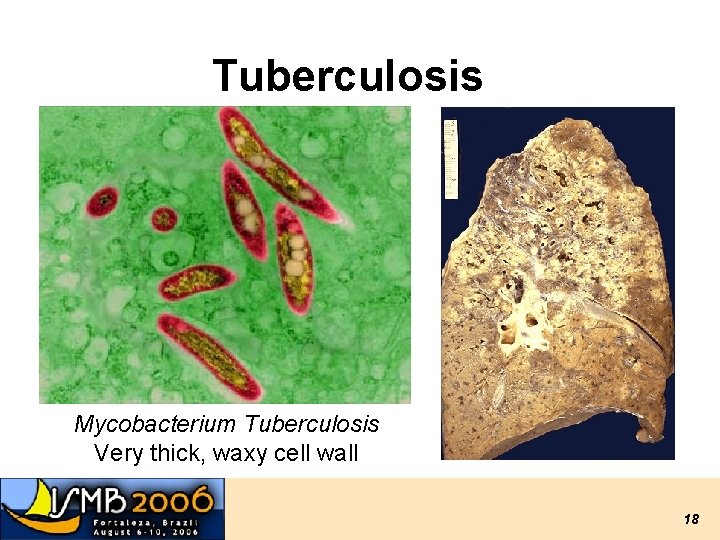
Tuberculosis Mycobacterium Tuberculosis Very thick, waxy cell wall 18

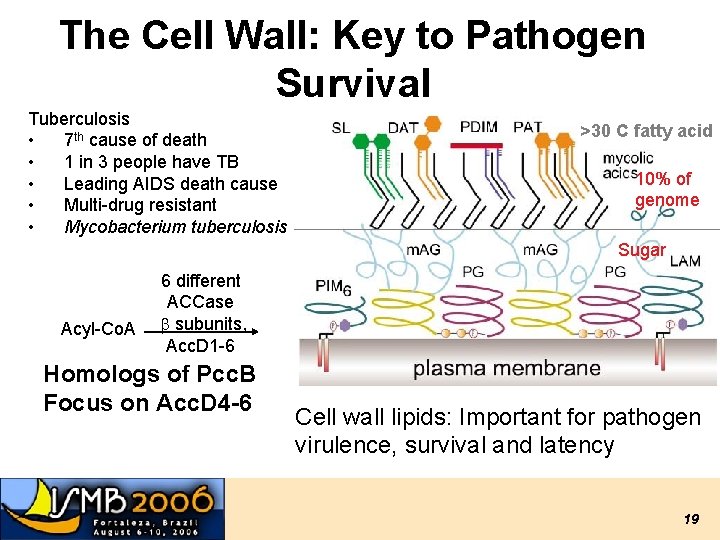
The Cell Wall: Key to Pathogen Survival Tuberculosis • 7 th cause of death • 1 in 3 people have TB • Leading AIDS death cause • Multi-drug resistant • Mycobacterium tuberculosis >30 C fatty acid 10% of genome Sugar Acyl-Co. A 6 different ACCase b subunits, Acc. D 1 -6 Homologs of Pcc. B Focus on Acc. D 4 -6 Cell wall lipids: Important for pathogen virulence, survival and latency 19

Tuberculosis (TB): An old foe 20

The White Death Frederic Chopin 1810 -1849 John Keats 1795 -1821 21

TB: still a real threat, because…. . Its ability to stay alive Multi-Drug Resistant (Super TB strain) 22

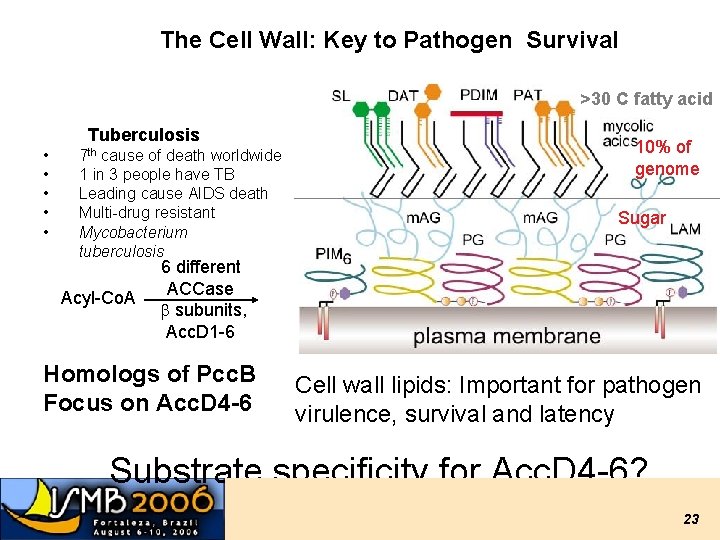
The Cell Wall: Key to Pathogen Survival >30 C fatty acid Tuberculosis • • • 7 th cause of death worldwide 1 in 3 people have TB Leading cause AIDS death Multi-drug resistant Mycobacterium tuberculosis Acyl-Co. A 10% of genome Sugar 6 different ACCase b subunits, Acc. D 1 -6 Homologs of Pcc. B Focus on Acc. D 4 -6 Cell wall lipids: Important for pathogen virulence, survival and latency Substrate specificity for Acc. D 4 -6? 23

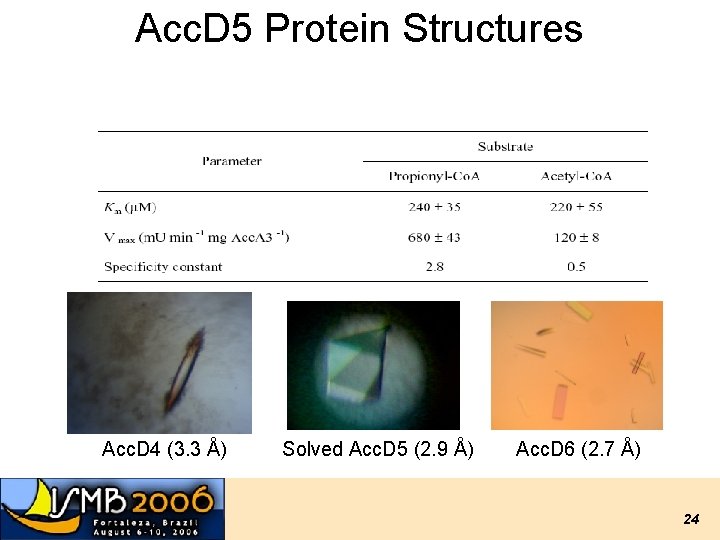
Acc. D 5 Protein Structures Acc. D 4 (3. 3 Å) Solved Acc. D 5 (2. 9 Å) Acc. D 6 (2. 7 Å) 24

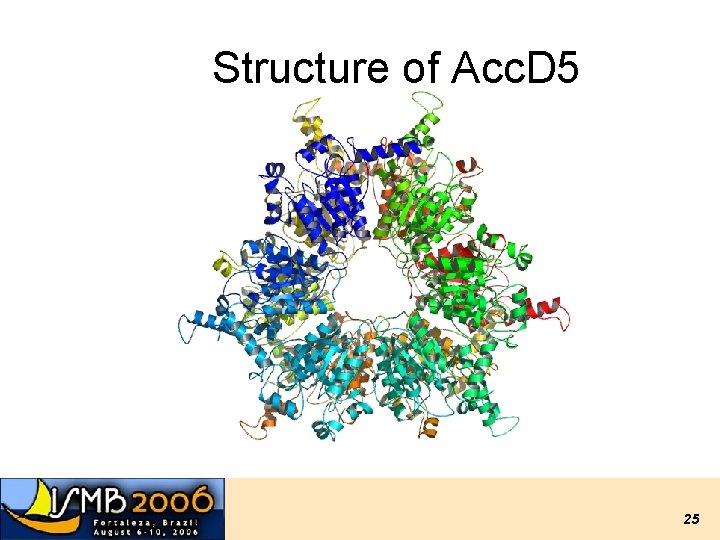
Structure of Acc. D 5 25

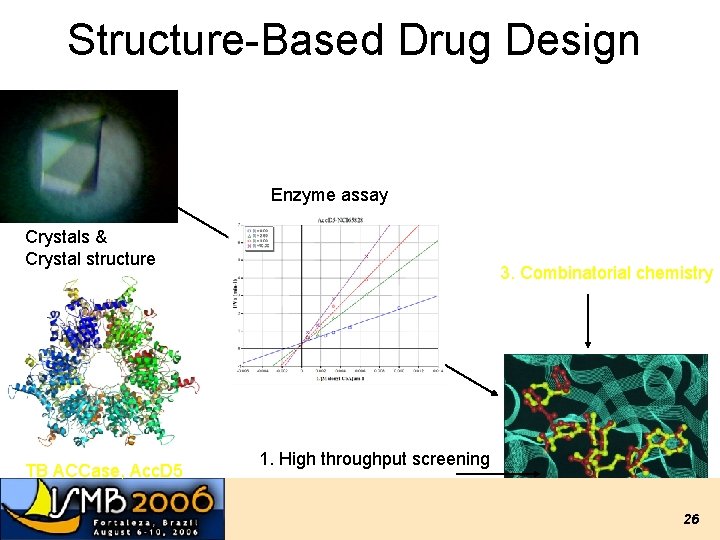
Structure-Based Drug Design Enzyme assay Crystals & Crystal structure TB ACCase, Acc. D 5 3. Combinatorial chemistry 1. High throughput screening 2. Virtual Screening Lead compound 26

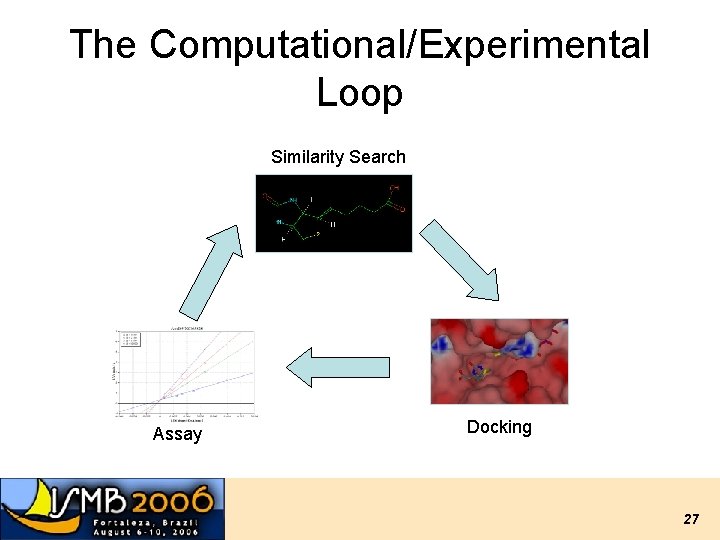
The Computational/Experimental Loop Similarity Search Assay Docking 27

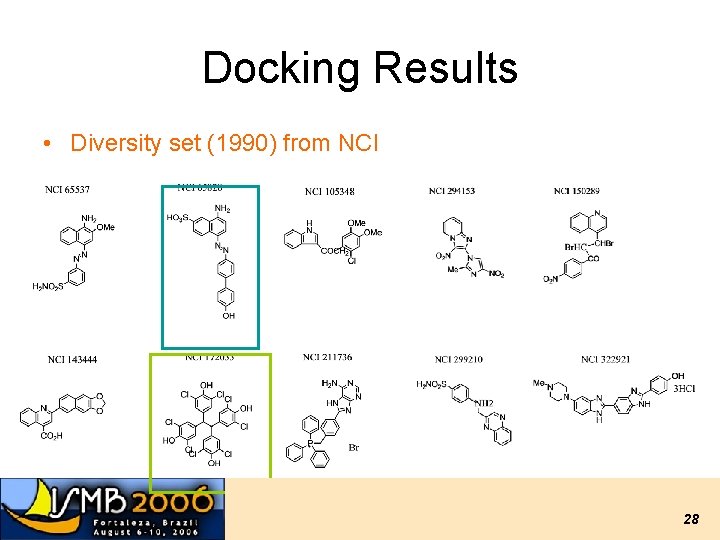
Docking Results • Diversity set (1990) from NCI 28

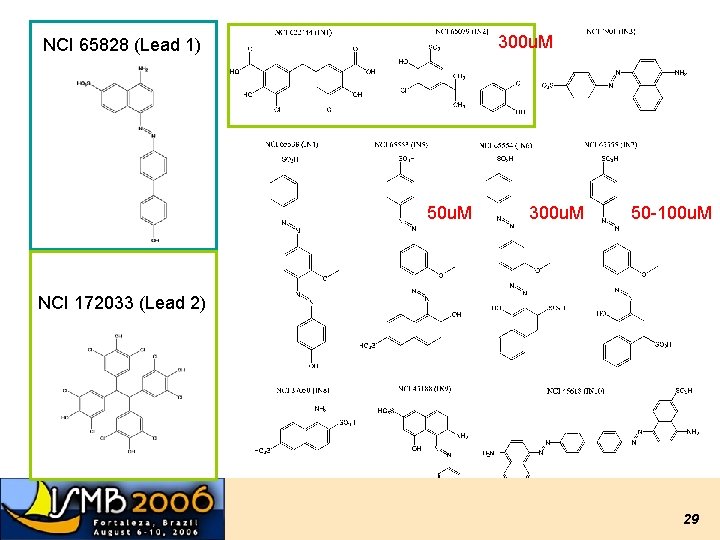
300 u. M NCI 65828 (Lead 1) 50 u. M 300 u. M 50 -100 u. M NCI 172033 (Lead 2) 29

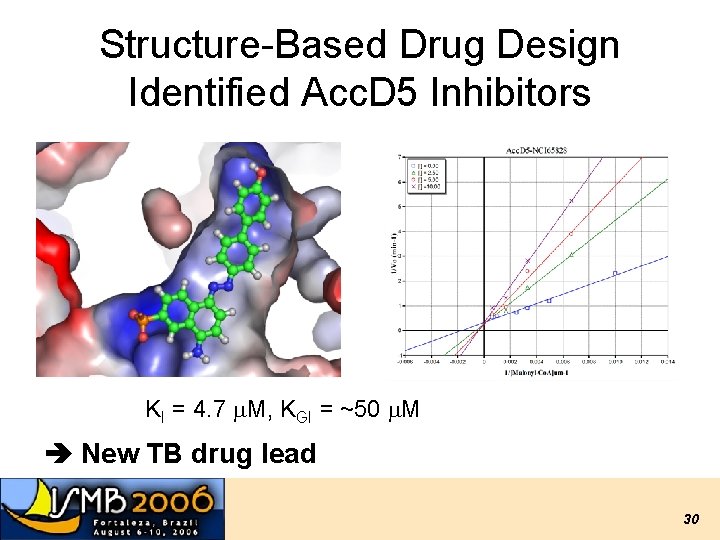
Structure-Based Drug Design Identified Acc. D 5 Inhibitors KI = 4. 7 m. M, KGI = ~50 m. M New TB drug lead T. Lin, M. Melgar, S. J. Swamidass, J. Purdon, T. Tseng, G. Gago, D. Kurth, P. Baldi, H. Gramajo, and S. Tsai. PNAS, 103, 9, 3072 -3077, (2006). US Patent pending. 30

Acknowledgements • Informatics – – – – – • Liva Ralaivola J. Chen S. J. Swamidass Yimeng Dou Peter Phung Jocelyne Bruand Chloe Azencott Alex Ksikes Ryan Allison Funding – – • Pharmacology – Daniele Piomelli • Chemistry – – – G. Weiss J. S. Nowick R. Chamberlin S. Tsai K. Shea NIH NSF Sun IGB 31

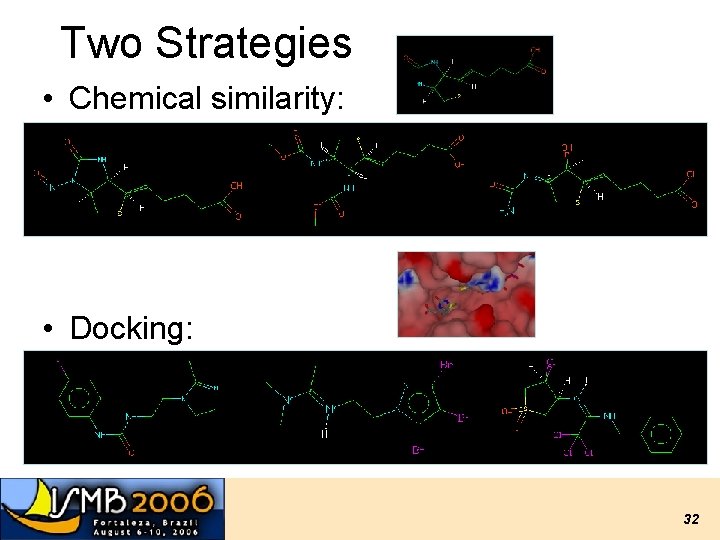
Two Strategies • Chemical similarity: • Docking: 32

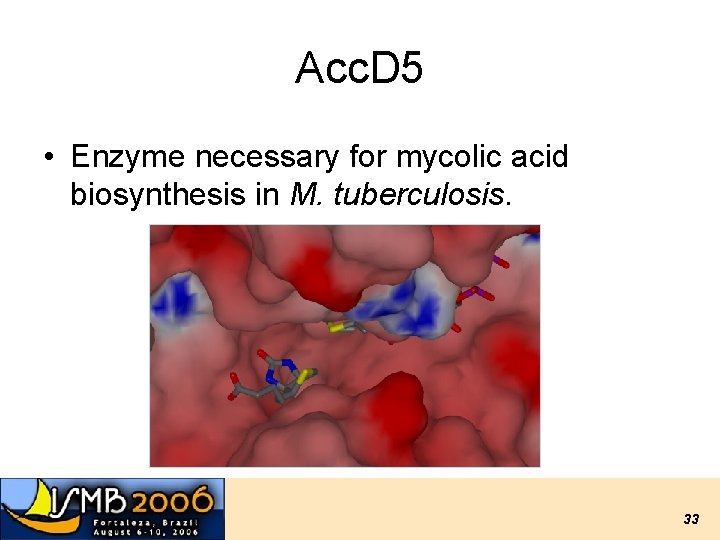
Acc. D 5 • Enzyme necessary for mycolic acid biosynthesis in M. tuberculosis. 33