7 Mechanisms of Mutation and DNA Repair 1










- Slides: 35

7 Mechanisms of Mutation and DNA Repair 1

Mutations • Spontaneous mutation: occurs in absence of mutagenic agent • Rate of mutation: probability of change in DNA sequence during a single generation • Induced mutation: caused by exposure to mutagenic agent=mutagen 2

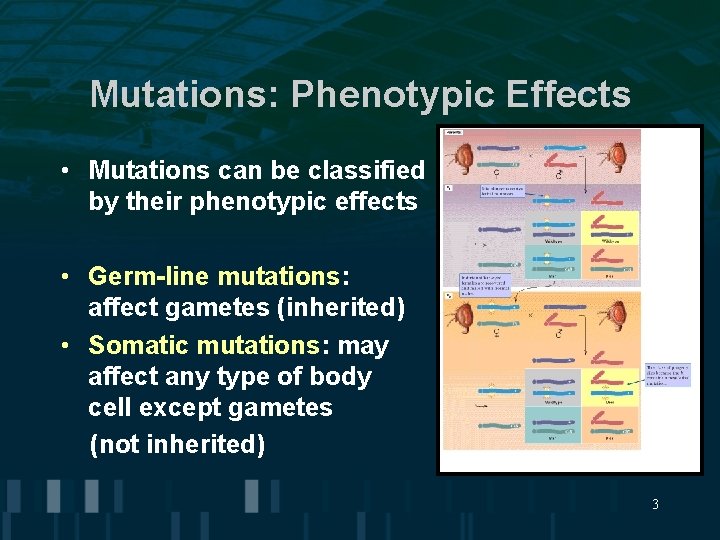
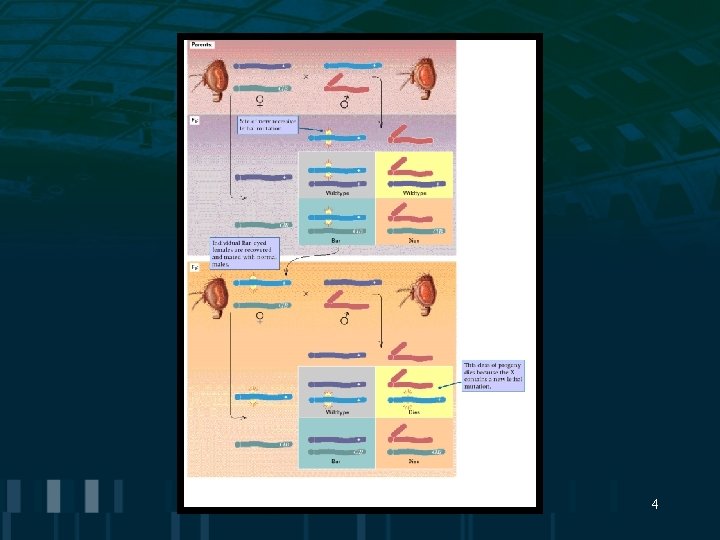
Mutations: Phenotypic Effects • Mutations can be classified by their phenotypic effects • Germ-line mutations: affect gametes (inherited) • Somatic mutations: may affect any type of body cell except gametes (not inherited) 3

4

Mutations • Conditional mutations: produce phenotypic changes under specific (restrictive) conditions but not others (permissive conditions) • Temperature-sensitive mutations: conditional mutation whose expression depends on temperature 5

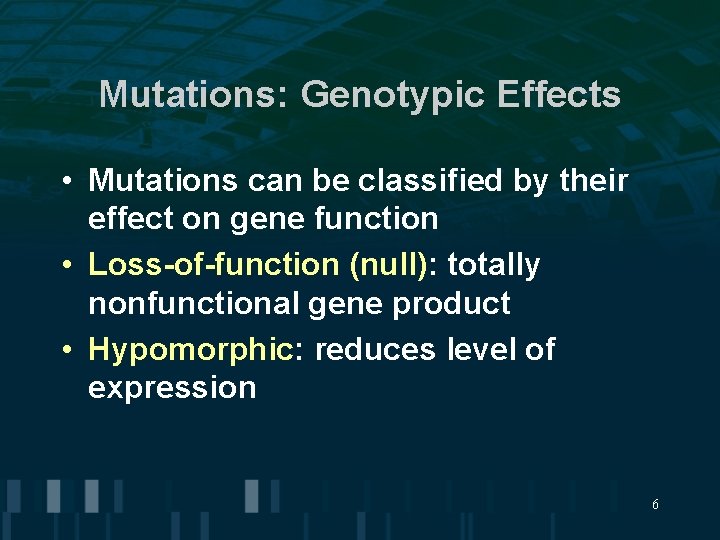
Mutations: Genotypic Effects • Mutations can be classified by their effect on gene function • Loss-of-function (null): totally nonfunctional gene product • Hypomorphic: reduces level of expression 6

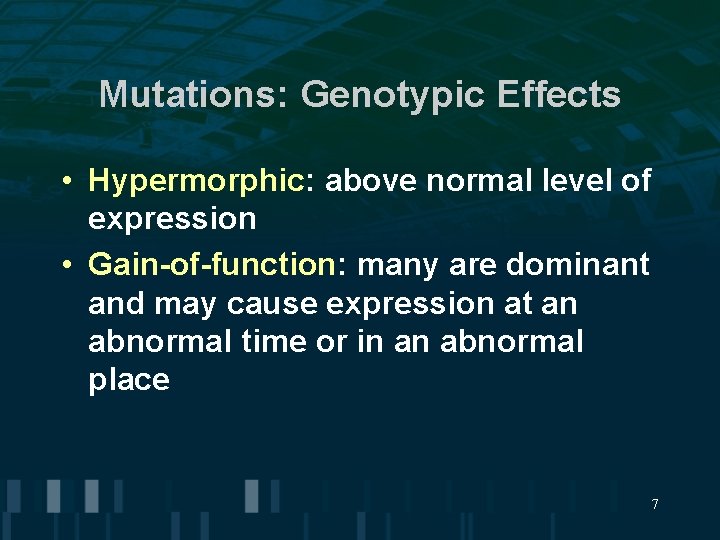
Mutations: Genotypic Effects • Hypermorphic: above normal level of expression • Gain-of-function: many are dominant and may cause expression at an abnormal time or in an abnormal place 7

Molecular Basis of Mutations result from changes in the base sequence of DNA: • Base substitutions -one pair of of DNA nucleotides is replaced by another pair : -Transition mutations- a purine is substituted for a purine or a pyrimidine is substituted for a pyrimidine 8

9

Molecular Basis of Mutation -Transversion mutations- a purine replaces a pyrimidine or vice versa • Base substitutions are point mutations which alter one DNA base pair without adding or deleting any base pairs • Point mutations may affect gene expression in several ways 10

Point Mutations Types of point mutations: • Silent substitutions are base substitutions which do not alter the amino acid composition of the protein encoded by a gene: -silent mutations may affect the noncoding portion of a gene or may occur in the coding portion but may not alter codon usage 11

Point Mutations • Missense mutations change a single amino acid as a result of a change in codon specification: -missense mutations can have serious consequences on the biological properties of a protein - sickle cell anemia results from a single amino acid substitution in hemoglobin which alters its structure 12

Point Mutations Point mutations can also alter signals used to regulate gene expression: • Promoter mutations may block transcription • Splice site mutations may block splicing or create new splice signals • Nonsense mutations change a codon to a stop codon which results in a premature termination of translation 13

Insertions and Deletions • Insertions add one or more nucleotide pairs to DNA sequence • Deletions remove one or more nucleotide pairs from DNA sequence • Insertions or deletions involving a multiple of 3 DNA base pairs = in-frame since they do not alter the reading frame of the genetic code 14

Insertions and Deletions • Insertions or deletions which involve a non-multiple of 3 DNA base pairs = frameshift mutations since they alter the codon translation reading frame • Large deletions may remove genes-no gene product is made • Insertions can result from gene amplification which can result in the overproduction of gene products 15

Insertions and Deletions • Deletion mutations in the dystrophin gene cause muscular dystrophy • Gene amplifications are often observed in human malignancies • Insertion and deletion mutations may result from unequal crossing-over during recombination or replication slippage during replication of simple tandem repeat sequences 16

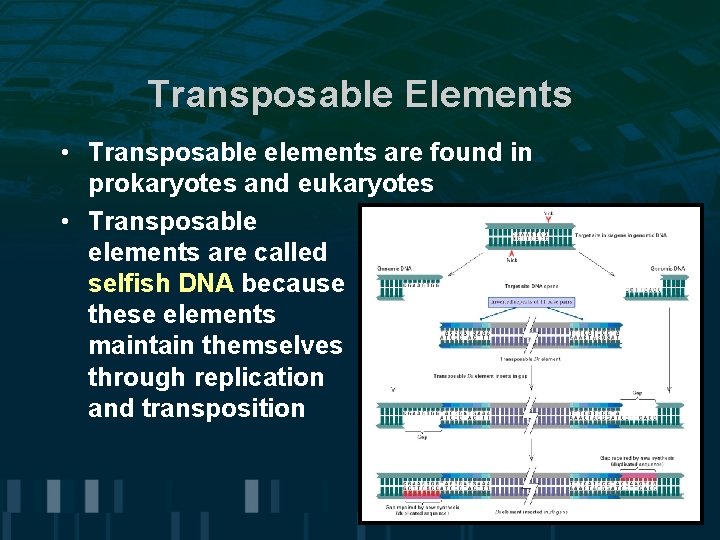
Transposable Elements • Transposable elements are found in prokaryotes and eukaryotes • Transposable elements are called selfish DNA because these elements maintain themselves through replication and transposition

18

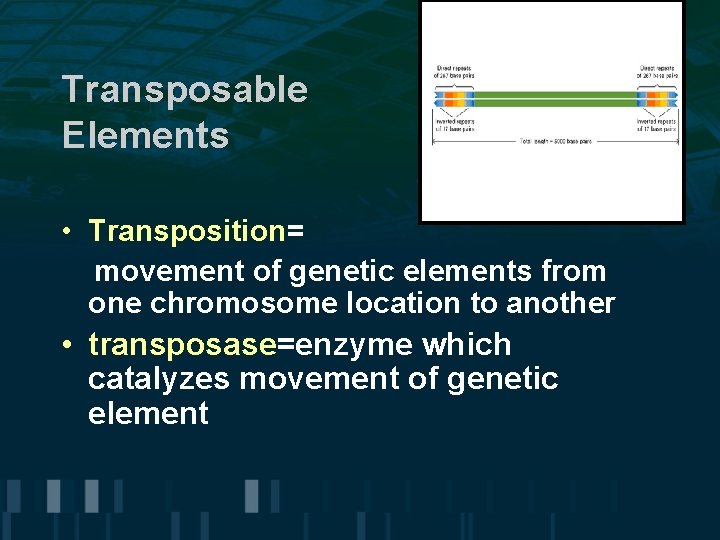
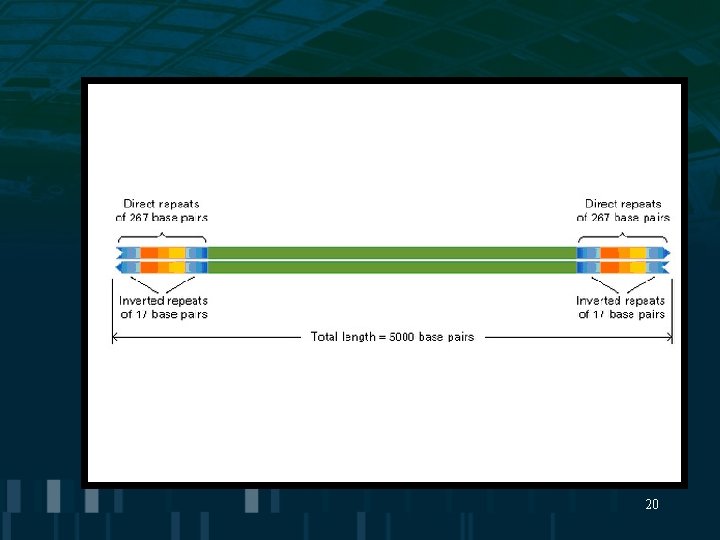
Transposable Elements • Transposition= movement of genetic elements from one chromosome location to another • transposase=enzyme which catalyzes movement of genetic element

20

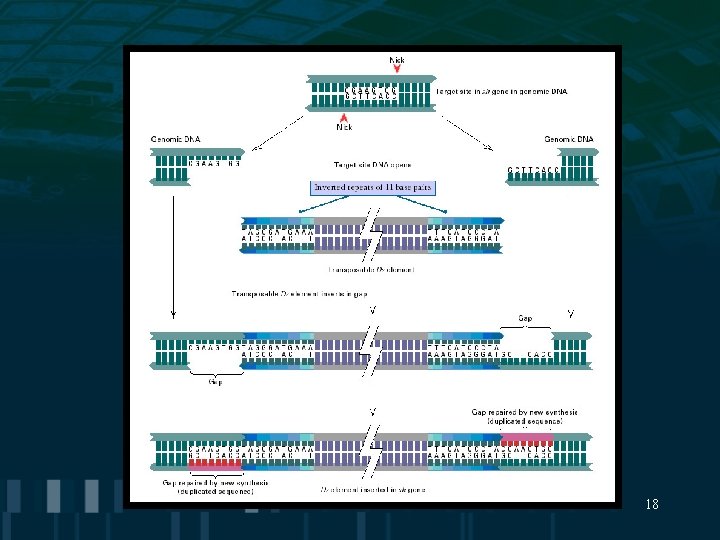
Transposable Elements • Steps in transposition: -transposase binds to terminal inverted repeat sequence -enzymatic cleavage results in transfer to different chromosomal site -insertion site is random and involves duplication of 2 -12 base pairs • Transposable elements cause mutations by inactivating genes at sites of insertion

Transposable Elements • Reverse transcriptase: enzyme using RNA transcript as a template for a DNA daughter strand • LTR retrotransposons: long terminal repeats • Non-LTR retrotransposons: no terminal repeats – LINE and SINE: most abundant transposable elements in mammalian genomes 22

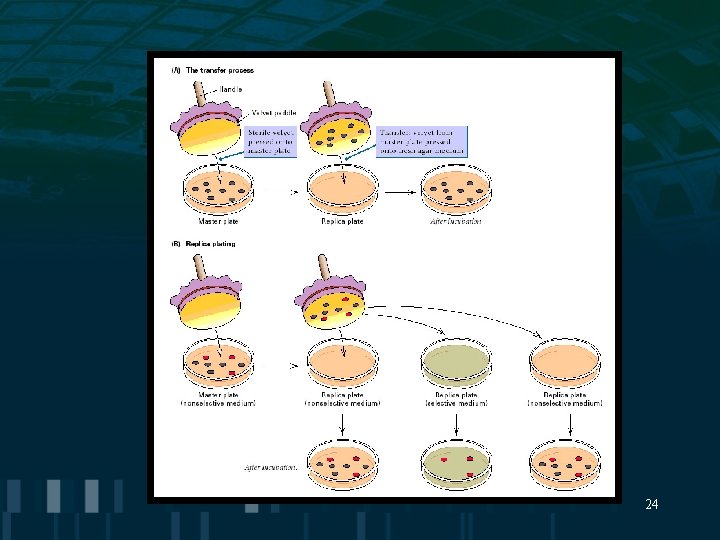
Spontaneous Mutations Lederberg’s replica plating: • bacterial colonies are transferred to velvet pad and from pad to new plate to test for the frequency of phage resistant colonies in a • population 23

24

Mutation Hot Spots Mutation hot spots have a higher mutation rate than most DNA: • Cytosine deamination to uracil is often detected at hot spots • Sites of cytosine methylation result in deamination which converts 5 methylcytosine to thymine • Both mutations result in GC to AT transitions 25

Mutation Hot Spots • Cytosine deamination can be repaired by DNA uracil glycosylase which recognizes the incorrect GU base pair and removes uracil • AP endonuclease then removes the ribose sugar • Single-strand gap is repaired by DNA polymerases and nick is sealed by ligase 26

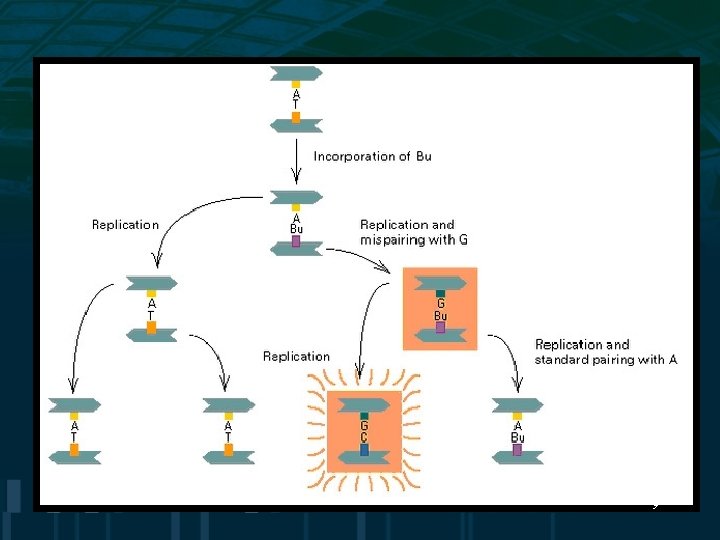
Induced Mutations • Base analogs such as 5 -bromouracil may be incorporated into DNA during replication instead of thymine and pairs with guanine resulting in AT to GC transition • Nucleotide analogs can inhibit DNA replication 27

Chemical Mutagenesis • Nitrous acid converts amino groups to keto groups altering the base pairing properties of the bases to produce transition mutations • Alkylating agents add alkyl groups to bases resulting in transition mutations or depurination = loss of guanine 28

Radiation Mutagenesis • Ultraviolet radiation (UV) causes adjacent thymines to become covalently linked = pyrimidine dimers • Ionizing radiation causes formation of free radicals, highly reactive ions which can damage DNA producing serious mutagenic effects 29

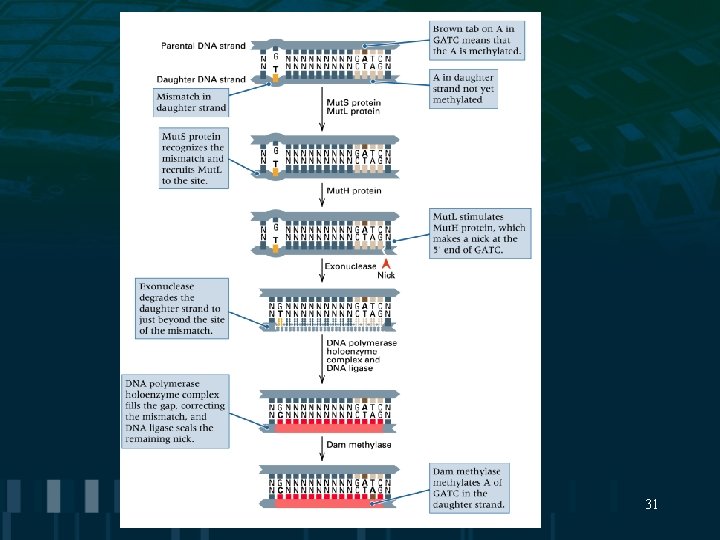
DNA Repair Mechanisms • Mismatch Repair consists of the excision of a segment of DNA that contains a base mismatch followed by repair synthesis • Photoreactivation repairs UV- induced pyrimidine dimers by breaking the covalent linkage between the thymine bases 30

31

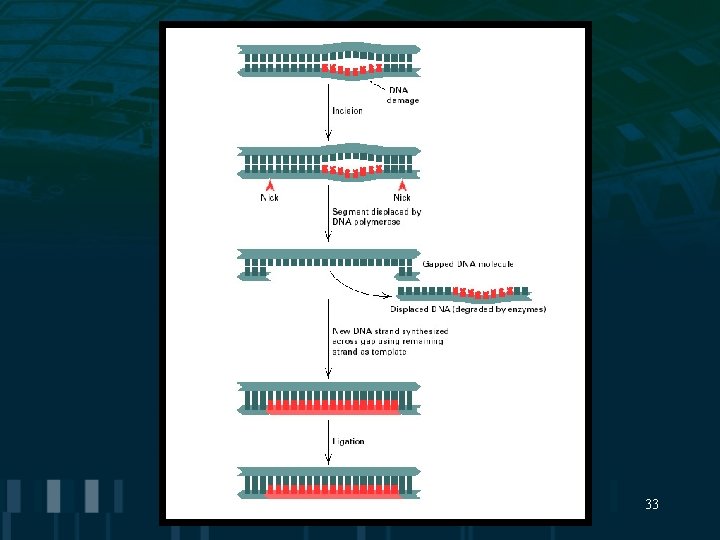
Excision Repair • Excision repair is a multistep process in which a segment of damaged DNA is removed and replaced by Resynthesis using the undamaged strand as a template 32

33

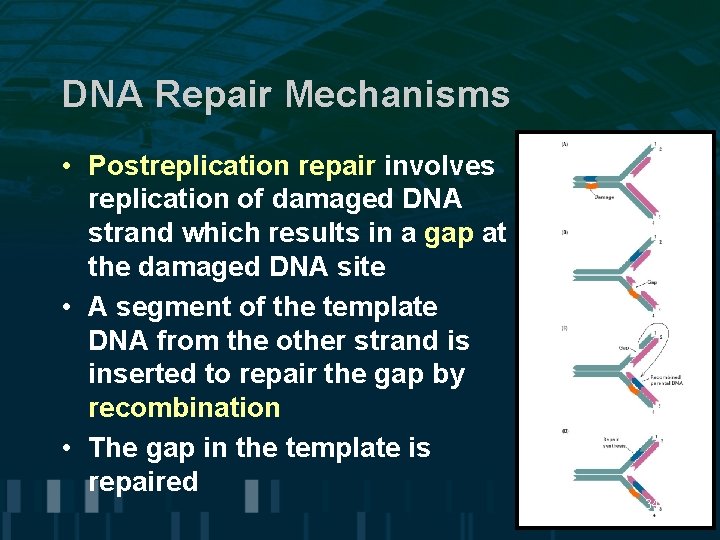
DNA Repair Mechanisms • Postreplication repair involves replication of damaged DNA strand which results in a gap at the damaged DNA site • A segment of the template DNA from the other strand is inserted to repair the gap by recombination • The gap in the template is repaired 34

35