3747 THOMAS THALAMUS OPTIMIZED MULTIATLAS SEGMENTATION Jason Su


- Slides: 10

#3747 THOMAS: THALAMUS OPTIMIZED MULTI-ATLAS SEGMENTATION Jason Su 1, 2, Thomas Tourdias 3, Manojkumar Saranathan 2, and Brian K. Rutt 2 1 Department of Electrical Engineering, Stanford University, Stanford, California, USA 2 Department of Radiology, Stanford University, Stanford, California, USA 3 Department of Neuroradiology, Bordeaux University Hospital, Bordeaux, France Declaration of Conflict of Interest or Relationship Research partially supported by a grant from GE Healthcare.

THOMAS: THALAMUS OPTIMIZED MULTI-ATLAS SEGMENTATION #3747 Background White matter nulled MPRAGE (WMn. MPRAGE) at 7 T reveals details in the thalamus Enabled detailed delineation of 15 thalamic nuclei guided by the Morel atlas 1, 2 1 Tourdias 2 Niemann et al. Neuroimage. 2013 Sep 7; 84 C: 534 -545. et al. Neuroimage. 2000 Dec; 12(6): 601 -16. Manual segmentation of thalamic nuclei using a WMn. MPRAGE acquisition.

THOMAS: THALAMUS OPTIMIZED MULTI-ATLAS SEGMENTATION #3747 Background Segmentation of thalamic nuclei is an important task • Diseases and disorders like Multiple Sclerosis and Essential Tremor can cause atrophy in the thalamus • Helpful for surgical guidance, especially MRg. FUS However, manual delineation takes up to a day. Can we make it automatic? Manual segmentation of thalamic nuclei using a WMn. MPRAGE acquisition.

THOMAS: THALAMUS OPTIMIZED MULTI-ATLAS SEGMENTATION #3747 Outline Automatically segment the thalamus and its nuclei using a library of manual-defined atlases Impact: Optimize for thalamic segmentation Enables large-scale study of thalamic atrophy and can aid in surgical planning. Assess its accuracy against manual delineation

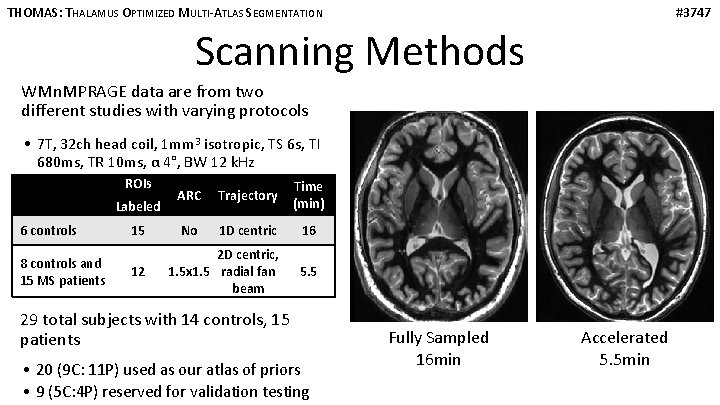
THOMAS: THALAMUS OPTIMIZED MULTI-ATLAS SEGMENTATION #3747 Scanning Methods WMn. MPRAGE data are from two different studies with varying protocols • 7 T, 32 ch head coil, 1 mm 3 isotropic, TS 6 s, TI 680 ms, TR 10 ms, α 4°, BW 12 k. Hz ROIs Labeled 6 controls 8 controls and 15 MS patients 15 12 ARC Trajectory Time (min) No 1 D centric 16 2 D centric, 1. 5 x 1. 5 radial fan beam 5. 5 29 total subjects with 14 controls, 15 patients • 20 (9 C: 11 P) used as our atlas of priors • 9 (5 C: 4 P) reserved for validation testing Fully Sampled 16 min Accelerated 5. 5 min

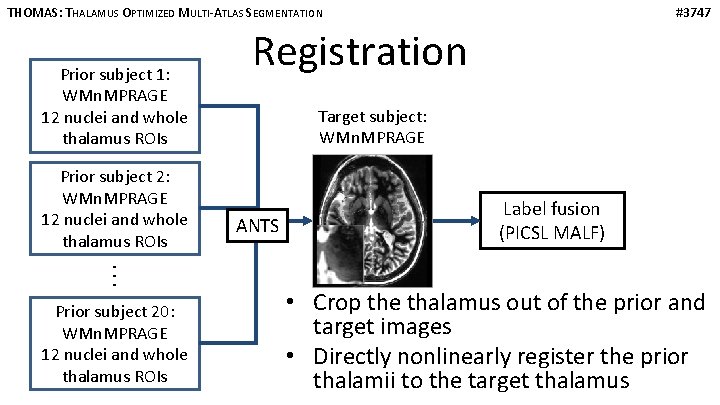
THOMAS: THALAMUS OPTIMIZED MULTI-ATLAS SEGMENTATION Prior subject 1: WMn. MPRAGE 12 nuclei and whole thalamus ROIs Prior subject 2: WMn. MPRAGE 12 nuclei and whole thalamus ROIs · · · Prior subject 20: WMn. MPRAGE 12 nuclei and whole thalamus ROIs #3747 Registration Target subject: WMn. MPRAGE ANTS Label fusion (PICSL MALF) • Crop the thalamus out of the prior and target images • Directly nonlinearly register the prior thalamii to the target thalamus

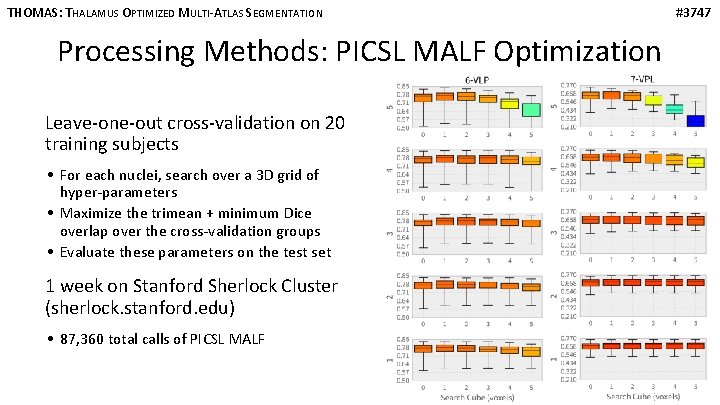
THOMAS: THALAMUS OPTIMIZED MULTI-ATLAS SEGMENTATION Processing Methods: PICSL MALF Optimization Leave-one-out cross-validation on 20 training subjects • For each nuclei, search over a 3 D grid of hyper-parameters • Maximize the trimean + minimum Dice overlap over the cross-validation groups • Evaluate these parameters on the test set 1 week on Stanford Sherlock Cluster (sherlock. stanford. edu) • 87, 360 total calls of PICSL MALF #3747

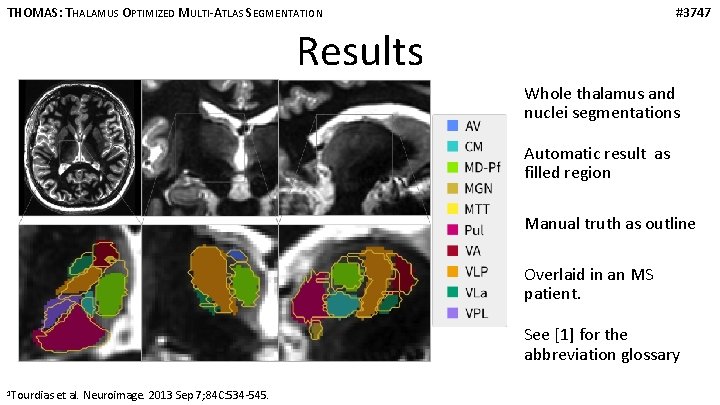
THOMAS: THALAMUS OPTIMIZED MULTI-ATLAS SEGMENTATION #3747 Results Whole thalamus and nuclei segmentations Automatic result as filled region Manual truth as outline Overlaid in an MS patient. See [1] for the abbreviation glossary 1 Tourdias et al. Neuroimage. 2013 Sep 7; 84 C: 534 -545.

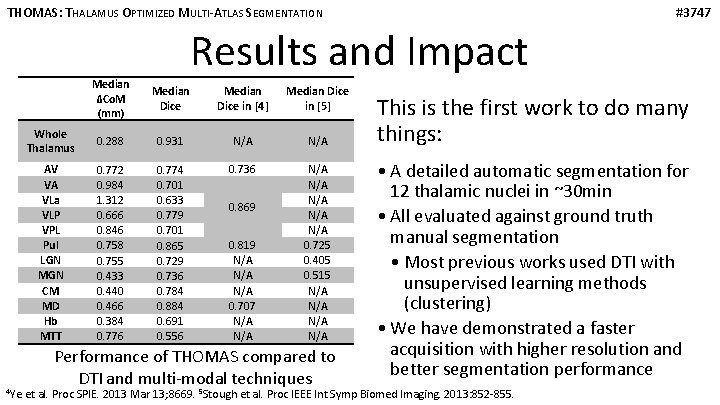
THOMAS: THALAMUS OPTIMIZED MULTI-ATLAS SEGMENTATION #3747 Results and Impact 4 Ye Median ΔCo. M (mm) Median Dice in [4] Median Dice in [5] Whole Thalamus 0. 288 0. 931 N/A AV VA VLa VLP VPL Pul LGN MGN CM MD Hb MTT 0. 772 0. 984 1. 312 0. 666 0. 846 0. 758 0. 755 0. 433 0. 440 0. 466 0. 384 0. 776 0. 774 0. 701 0. 633 0. 779 0. 701 0. 865 0. 729 0. 736 0. 784 0. 884 0. 691 0. 556 0. 736 N/A N/A N/A 0. 725 0. 405 0. 515 N/A N/A 0. 869 0. 819 N/A N/A 0. 707 N/A Performance of THOMAS compared to DTI and multi-modal techniques This is the first work to do many things: • A detailed automatic segmentation for 12 thalamic nuclei in ~30 min • All evaluated against ground truth manual segmentation • Most previous works used DTI with unsupervised learning methods (clustering) • We have demonstrated a faster acquisition with higher resolution and better segmentation performance et al. Proc SPIE. 2013 Mar 13; 8669. 5 Stough et al. Proc IEEE Int Symp Biomed Imaging. 2013: 852 -855.

THOMAS: THALAMUS OPTIMIZED MULTI-ATLAS SEGMENTATION Future Work Exploring several other registration architectures to improve robustness 6 Preliminary success with 3 T images using whole brain registration (4 -5 hours) Eliminated interpolation errors by using vector ROI models 6 Pipitone et al. Neuroimage. 2014 Nov 1; 101: 494 -512. #3747