3 DEM DAS Extending DAS to 3 DEM





- Slides: 15

3 D-EM DAS Extending DAS to 3 D-EM and Fitting http: //biocomp. cnb. uam. es/das/ jrmacias@cnb. uam. es 1 2007/02/26

Why 3 D-EM ? jrmacias@cnb. uam. es Three-Dimensional Electron Microscopy Single particle EM. 2 2007/02/26

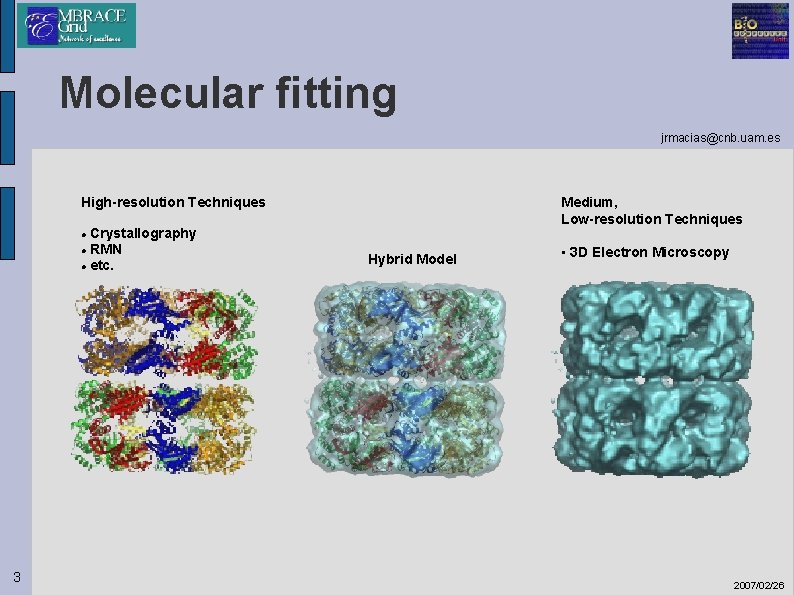
Molecular fitting jrmacias@cnb. uam. es High-resolution Techniques Crystallography RMN etc. Medium, Low-resolution Techniques 3 Hybrid Model • 3 D Electron Microscopy 2007/02/26

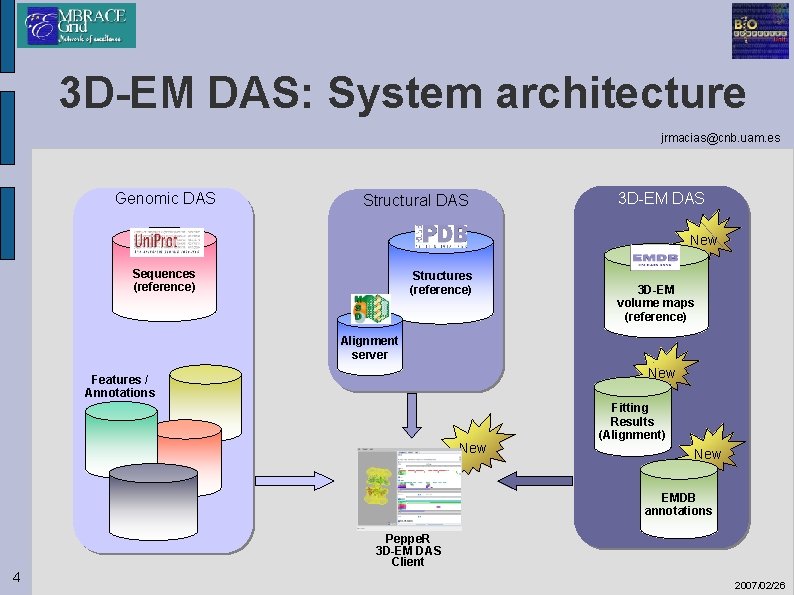
3 D-EM DAS: System architecture jrmacias@cnb. uam. es Genomic DAS Structural DAS 3 D-EM DAS New Sequences (reference) Structures (reference) 3 D-EM volume maps (reference) Alignment server New Features / Annotations New Fitting Results (Alignment) New EMDB annotations Peppe. R 3 D-EM DAS Client 4 2007/02/26

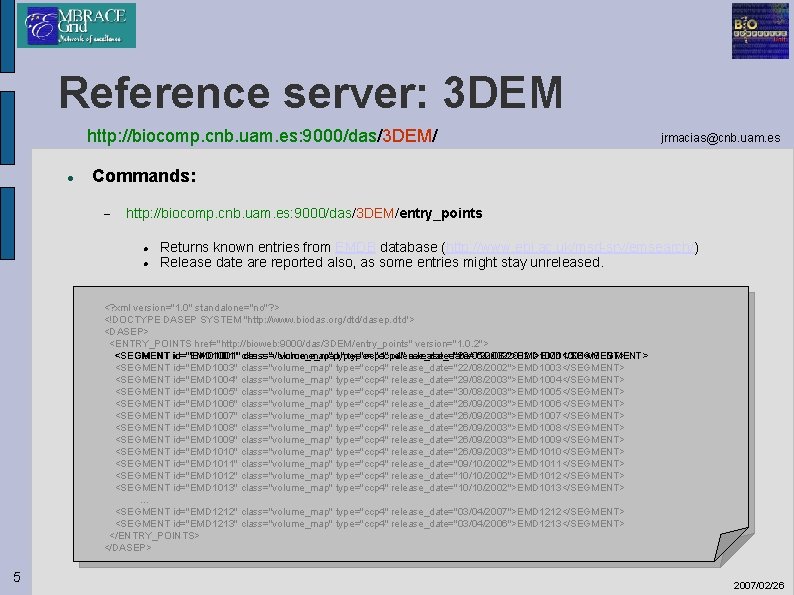
Reference server: 3 DEM http: //biocomp. cnb. uam. es: 9000/das/3 DEM/ jrmacias@cnb. uam. es Commands: http: //biocomp. cnb. uam. es: 9000/das/3 DEM/entry_points Returns known entries from EMDB database (http: //www. ebi. ac. uk/msd-srv/emsearch/) Release date are reported also, as some entries might stay unreleased. <? xml version="1. 0" standalone="no"? > <!DOCTYPE DASEP SYSTEM "http: //www. biodas. org/dtd/dasep. dtd"> <DASEP> <ENTRY_POINTS href="http: //bioweb: 9000/das/3 DEM/entry_points" version="1. 0. 2"> <SEGMENT id="EMD 1001" class="volume_map" type="ccp 4" release_date="20/06/2002">EMD 1001</SEGMENT> <SEGMENT id="EMD 1003" class="volume_map" type="ccp 4" release_date="22/08/2002">EMD 1003</SEGMENT> <SEGMENT id="EMD 1004" class="volume_map" type="ccp 4" release_date="29/08/2003">EMD 1004</SEGMENT> <SEGMENT id="EMD 1005" class="volume_map" type="ccp 4" release_date="30/08/2003">EMD 1005</SEGMENT> <SEGMENT id="EMD 1006" class="volume_map" type="ccp 4" release_date="26/09/2003">EMD 1006</SEGMENT> <SEGMENT id="EMD 1007" class="volume_map" type="ccp 4" release_date="26/09/2003">EMD 1007</SEGMENT> <SEGMENT id="EMD 1008" class="volume_map" type="ccp 4" release_date="26/09/2003">EMD 1008</SEGMENT> <SEGMENT id="EMD 1009" class="volume_map" type="ccp 4" release_date="26/09/2003">EMD 1009</SEGMENT> <SEGMENT id="EMD 1010" class="volume_map" type="ccp 4" release_date="26/09/2003">EMD 1010</SEGMENT> <SEGMENT id="EMD 1011" class="volume_map" type="ccp 4" release_date="09/10/2002">EMD 1011</SEGMENT> <SEGMENT id="EMD 1012" class="volume_map" type="ccp 4" release_date="10/10/2002">EMD 1012</SEGMENT> <SEGMENT id="EMD 1013" class="volume_map" type="ccp 4" release_date="10/10/2002">EMD 1013</SEGMENT> … <SEGMENT id="EMD 1212" class="volume_map" type="ccp 4" release_date="03/04/2007">EMD 1212</SEGMENT> <SEGMENT id="EMD 1213" class="volume_map" type="ccp 4" release_date="03/04/2006">EMD 1213</SEGMENT> </ENTRY_POINTS> </DASEP> 5 2007/02/26

Reference server: 3 DEM http: //biocomp. cnb. uam. es: 9000/das/3 DEM/ jrmacias@cnb. uam. es Commands: New command: http: //biocomp. cnb. uam. es: 9000/das/3 DEM/volmap? query=EMD 1017 Returns the volume map (a link, actually) for the entry. Equivalent to dna or sequence queries in other DAS systems. New xml document definition: Equivalent to DASSEQUENCE <? xml version="1. 0" standalone="yes"? > <!DOCTYPE DASVOLMAP SYSTEM "http: //biocomp. cnb. uam. es/das/dtd/dasvolmap. dtd"> <DASVOLMAP> <VOLMAP id="EMD 1017" class="volume_map" type="ccp 4" version="1. 0"> <VOLMAP id="EMD 1017" <LINK href="ftp: //ftp. ebi. ac. uk/pub/databases/emdb/structures/EMD-1017/map/emd_1017. map. gz"> ftp: //ftp. ebi. ac. uk/pub/databases/emdb/structures/EMD-1017/map/emd_1017. map. gz </LINK> </VOLMAP> <NOTE> NOTE: The LINK element points to the location where a 3 D-EM DAS client should retrive the data for the volume map (ussully through ftp) </NOTE> </DASVOLMAP> 6 2007/02/26

Alignment server: 3 DEM_Fitting http: //biocomp. cnb. uam. es: 9000/das/3 DEM_Fitting/ 7 jrmacias@cnb. uam. es An alignment server… http: //biocomp. cnb. uam. es: 9000/das/3 DEM_Fitting/entry_points Returns the list of known fitting experiments. Currently this list is manually “curated” and updated from different sources, mainly EMDB. http: //biocomp. cnb. uam. es: 9000/das/3 DEM_Fitting/alignment? query=ID Where ID can be a fitting, a EMDB or PDB identifier. …plus an annotation server: http: //biocomp. cnb. uam. es: 9000/das/3 DEM_Fitting/types Returns used annotation types/categories. http: //biocomp. cnb. uam. es: 9000/das/3 DEM_Fitting/features? segment=FFT 0014 Reports annotations about the fitting experiment conditions: Method and software used. Experimentalist notes. etc. 2007/02/26

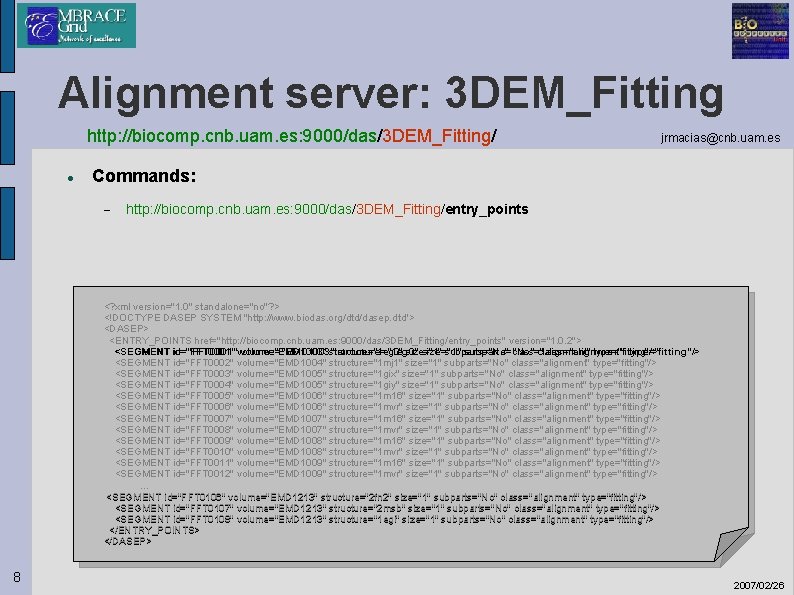
Alignment server: 3 DEM_Fitting http: //biocomp. cnb. uam. es: 9000/das/3 DEM_Fitting/ jrmacias@cnb. uam. es Commands: http: //biocomp. cnb. uam. es: 9000/das/3 DEM_Fitting/entry_points <? xml version="1. 0" standalone="no"? > <!DOCTYPE DASEP SYSTEM "http: //www. biodas. org/dtd/dasep. dtd"> <DASEP> <ENTRY_POINTS href="http: //biocomp. cnb. uam. es: 9000/das/3 DEM_Fitting/entry_points" version="1. 0. 2"> <SEGMENT id="FFT 0001" volume="EMD 1003" structure="1 eg 0" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0002" volume="EMD 1004" structure="1 mj 1" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0003" volume="EMD 1005" structure="1 gix" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0004" volume="EMD 1005" structure="1 giy" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0005" volume="EMD 1006" structure="1 m 16" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0006" volume="EMD 1006" structure="1 mvr" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0007" volume="EMD 1007" structure="1 m 16" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0008" volume="EMD 1007" structure="1 mvr" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0009" volume="EMD 1008" structure="1 m 16" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0010" volume="EMD 1008" structure="1 mvr" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0011" volume="EMD 1009" structure="1 m 16" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0012" volume="EMD 1009" structure="1 mvr" size="1" subparts="No" class="alignment" type="fitting"/> … <SEGMENT id="FFT 0106" volume="EMD 1213" structure="2 fn 2" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0107" volume="EMD 1213" structure="2 msb" size="1" subparts="No" class="alignment" type="fitting"/> <SEGMENT id="FFT 0108" volume="EMD 1213" structure="1 egi" size="1" subparts="No" class="alignment" type="fitting"/> </ENTRY_POINTS> </DASEP> 8 2007/02/26

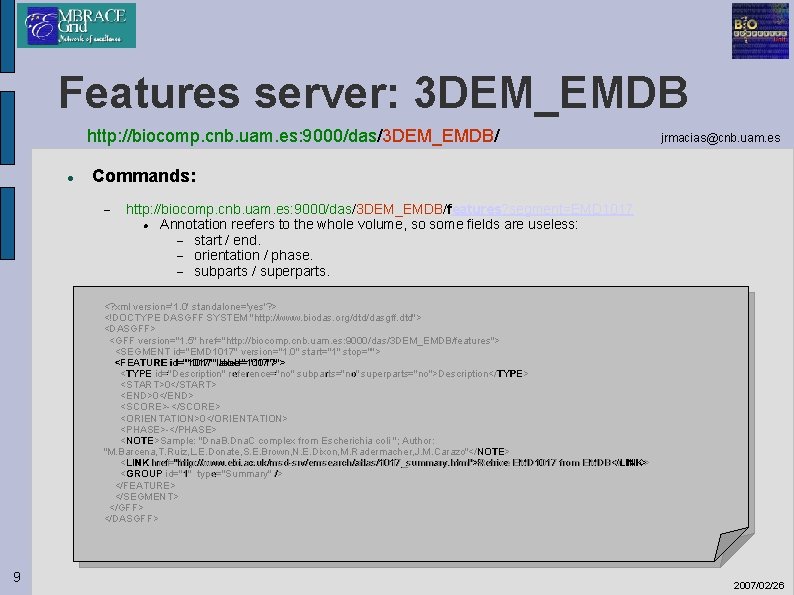
Features server: 3 DEM_EMDB http: //biocomp. cnb. uam. es: 9000/das/3 DEM_EMDB/ jrmacias@cnb. uam. es Commands: http: //biocomp. cnb. uam. es: 9000/das/3 DEM_EMDB/features? segment=EMD 1017 Annotation reefers to the whole volume, so some fields are useless: start / end. orientation / phase. subparts / superparts. <? xml version='1. 0' standalone='yes'? > <!DOCTYPE DASGFF SYSTEM "http: //www. biodas. org/dtd/dasgff. dtd"> <DASGFF> <GFF version="1. 5" href="http: //biocomp. cnb. uam. es: 9000/das/3 DEM_EMDB/features"> <SEGMENT id="EMD 1017" version="1. 0" start="1" stop=""> <FEATURE id="1017"label="1017"> <TYPE id="Description" reference="no" subparts="no" superparts="no">Description</TYPE> <START>0</START> <END>0</END> <SCORE>-</SCORE> <ORIENTATION>0</ORIENTATION> <PHASE>-</PHASE> <NOTE>Sample: "Dna. B. Dna. C complex from Escherichia coli "; Author: "M. Barcena, T. Ruiz, L. E. Donate, S. E. Brown, N. E. Dixon, M. Radermacher, J. M. Carazo"</NOTE> <LINK href="http: //www. ebi. ac. uk/msd-srv/emsearch/atlas/1017_summary. html">Retrive EMD 1017 from EMDB</LINK> <GROUP id="1" type="Summary" /> </FEATURE> </SEGMENT> </GFF> </DASGFF> 9 2007/02/26

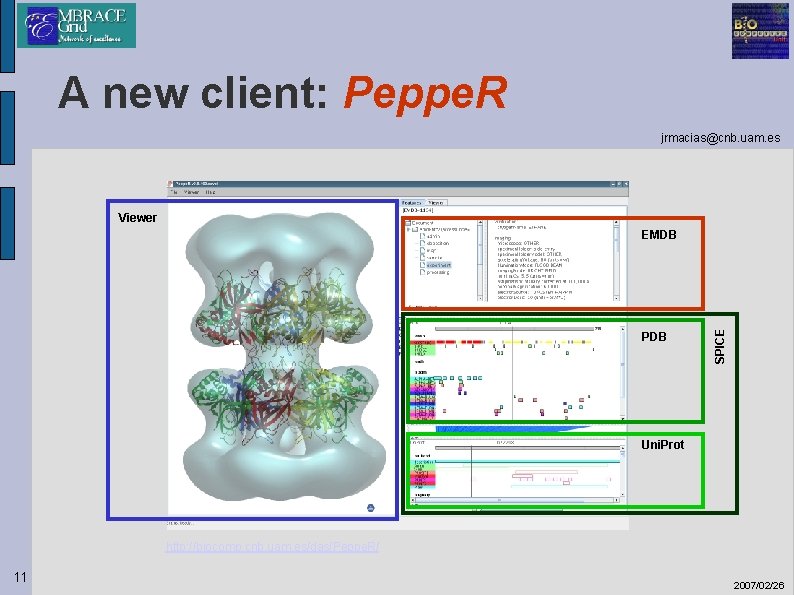
A new client: Peppe. R jrmacias@cnb. uam. es • Java implementation - Architecture independent • Windows • Unix / Linux • Mac Os • Starts as a stand-alone application or • Embedded in a web browser, as an applet. - Java Web Start deployment • Launched directly from a web link. • Easy version control and upgrade. • Uses Astex. Viewer to display maps + pdb structures together. • Features (annotations) panels from SPICE library. 10 2007/02/26

A new client: Peppe. R jrmacias@cnb. uam. es Viewer PDB SPICE EMDB Uni. Prot http: //biocomp. cnb. uam. es/das/Peppe. R/ 11 2007/02/26

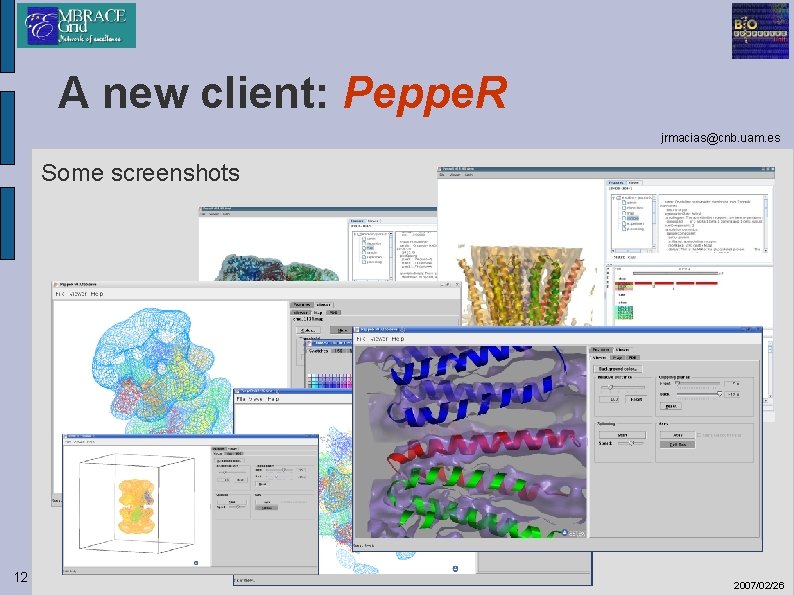
A new client: Peppe. R jrmacias@cnb. uam. es Some screenshots 12 2007/02/26

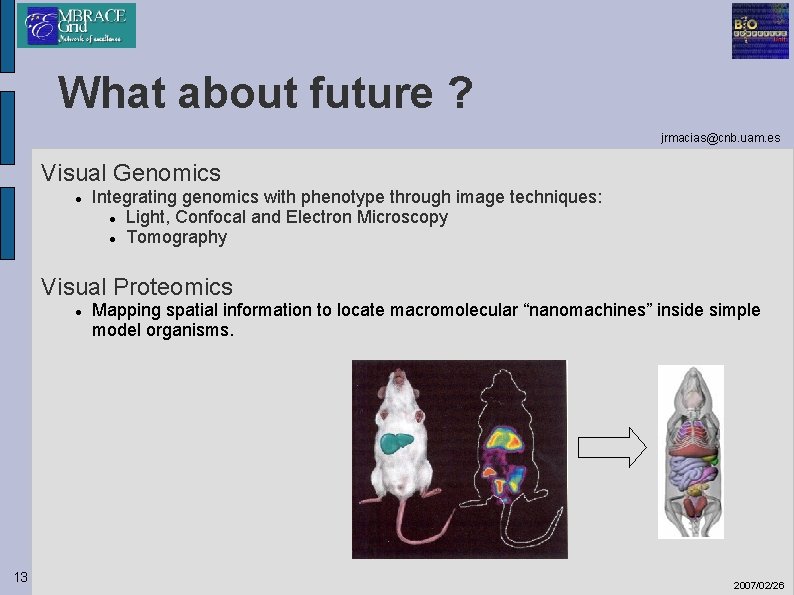
What about future ? jrmacias@cnb. uam. es Visual Genomics Integrating genomics with phenotype through image techniques: Light, Confocal and Electron Microscopy Tomography Visual Proteomics 13 Mapping spatial information to locate macromolecular “nanomachines” inside simple model organisms. 2007/02/26

What we’ll need ? jrmacias@cnb. uam. es NOW: Large data sets transfer : attachments? Avoid indirect access, through ftp SOON: 14 New types of annotations Ontology elements Phenotypic results Quantitative, continuous values Integration between different types of reference data Allowing more cross-references between different data sources. 2007/02/26

Acknowledgements: jrmacias@cnb. uam. es Centro Nacional de Biotecnologia Spanish National Center for Biology EMBRACE Grid Network of excellence Unidad de Biocomputación Bio. Computing Unit José Maria Carazo (head) 15 2007/02/26