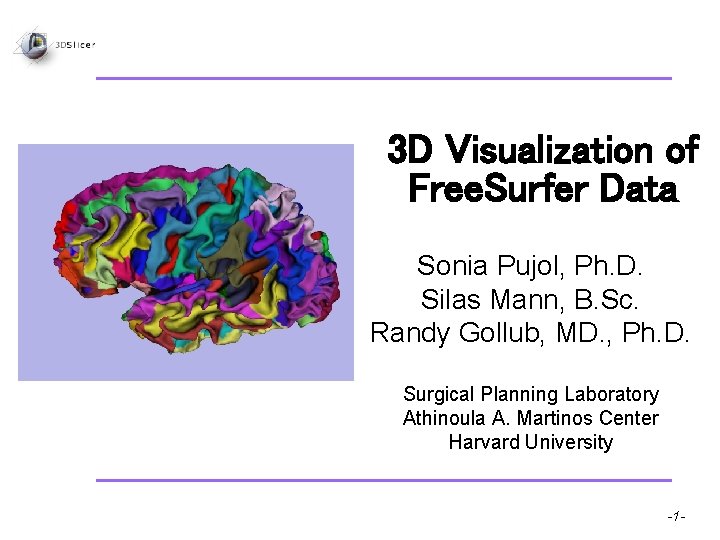
3 D Visualization of Free Surfer Data Sonia



- Slides: 59

3 D Visualization of Free. Surfer Data Sonia Pujol, Ph. D. Silas Mann, B. Sc. Randy Gollub, MD. , Ph. D. Surgical Planning Laboratory Athinoula A. Martinos Center Harvard University Pujol S et al. National Alliance for Medical Image Computing -1 -

Acknowledgements National Alliance for Medical Image Computing NIH U 54 EB 005149 Neuroimage Analysis Center NIH P 41 RR 013218 Morphometry Biomedical Informatics Research Network NIH U 24 RRO 21382 Surgical Planning Laboratory (BWH) Thanks to Nicole Aucoin Center for Functional Neuroimaging Technology NIH P 41 RR 14075 Pujol S et al. National Alliance for Medical Image Computing -2 -

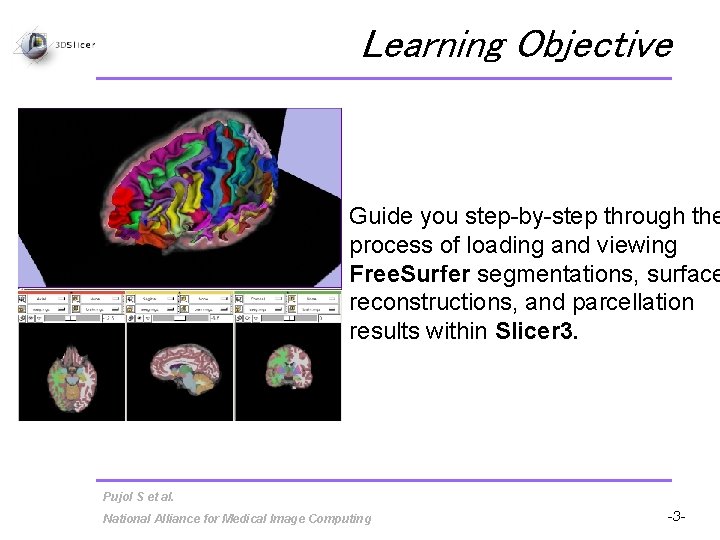
Learning Objective Guide you step-by-step through the process of loading and viewing Free. Surfer segmentations, surface reconstructions, and parcellation results within Slicer 3. Pujol S et al. National Alliance for Medical Image Computing -3 -

Prerequisites This tutorial assumes that you have completed the course Data Loading and Visualization. Tutorials for Slicer 3 are available on the Slicer 101 page: http: //www. na-mic. org/Wiki/index. php/Slicer 3. 2: Training Pujol S et al. National Alliance for Medical Image Computing -4 -

Prerequisites This tutorial assumes a working knowledge of how to use Free. Surfer to generate segmentation and surface files. Tutorials for Free. Surfer are available at the following location: http: //surfer. nmr. mgh. harvard. edu/fswiki/Tutorials/ Pujol S et al. National Alliance for Medical Image Computing -5 -

Materials This tutorial requires the installation of the Slicer 3 software and the tutorial dataset. These materials are available at the following locations: • Slicer 3 download page (Slicer 3. 2) http: //www. slicer. org/pages/Downloads Disclaimer: It is the responsibility of the user of Slicer to comply with both the terms of the license and with the applicable laws, regulations, and rules. Pujol S et al. National Alliance for Medical Image Computing -6 -

Materials This tutorial makes use of the same T 1 weighted image dataset (bert) that is used for the Free. Surfer tutorial available at the following location: http: //surfer. nmr. mgh. harvard. edu/fswiki/Fs. Tutorial If you already have the Free. Surfer subject ‘bert’ on your computer, then just download the file ‘slicer. Generic. Scene. mrml’ http: //www. na-mic. org/Wiki/index. php/Image: Slicer. Generic. Scene. mrml If you don’t have the Free. Surfer tutorial dataset known as ‘bert’ on your computer, then download the archive below: http: //www. na-mic. org/Wiki/index. php/Image: Free. Surfer. Data. tar. gz Pujol S et al. National Alliance for Medical Image Computing -7 -

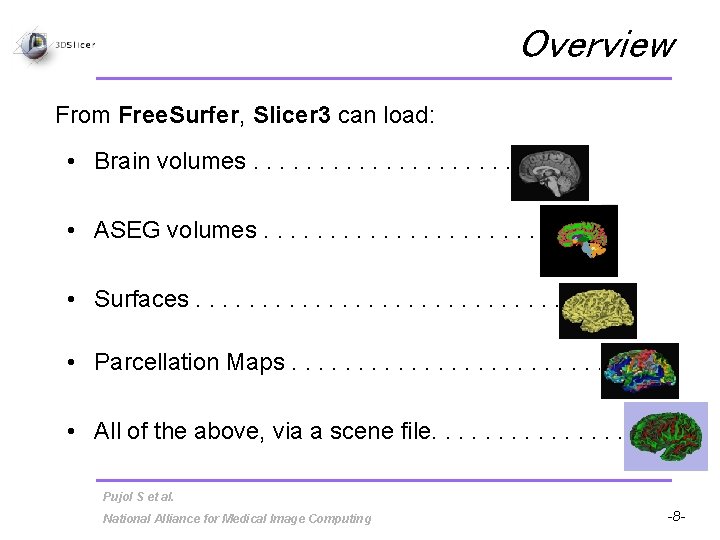
Overview From Free. Surfer, Slicer 3 can load: • Brain volumes. . . • ASEG volumes. . . . • Surfaces. . . . • Parcellation Maps. . . . • All of the above, via a scene file. . . . Pujol S et al. National Alliance for Medical Image Computing -8 -

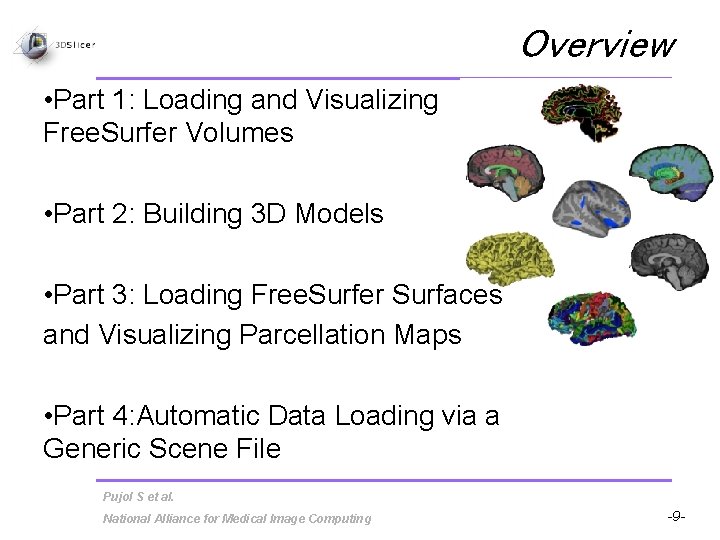
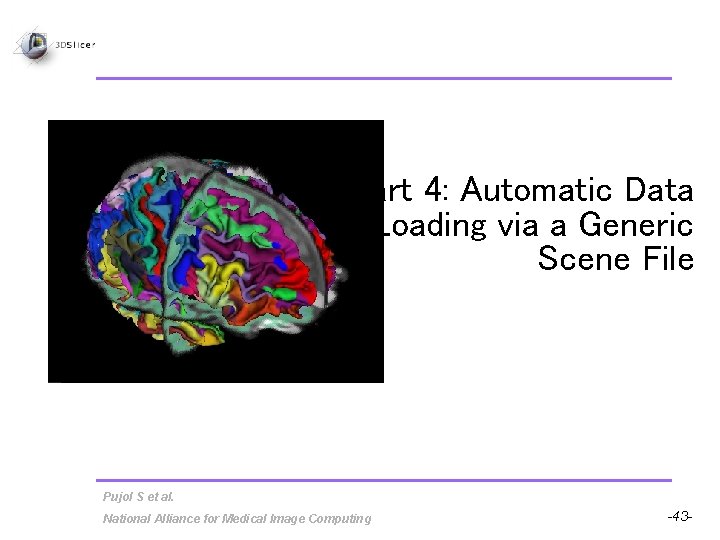
Overview • Part 1: Loading and Visualizing Free. Surfer Volumes • Part 2: Building 3 D Models • Part 3: Loading Free. Surfer Surfaces and Visualizing Parcellation Maps • Part 4: Automatic Data Loading via a Generic Scene File Pujol S et al. National Alliance for Medical Image Computing -9 -

Part 1: Loading and Visualizing Free. Surfer Volumes Pujol S et al. National Alliance for Medical Image Computing -10 -

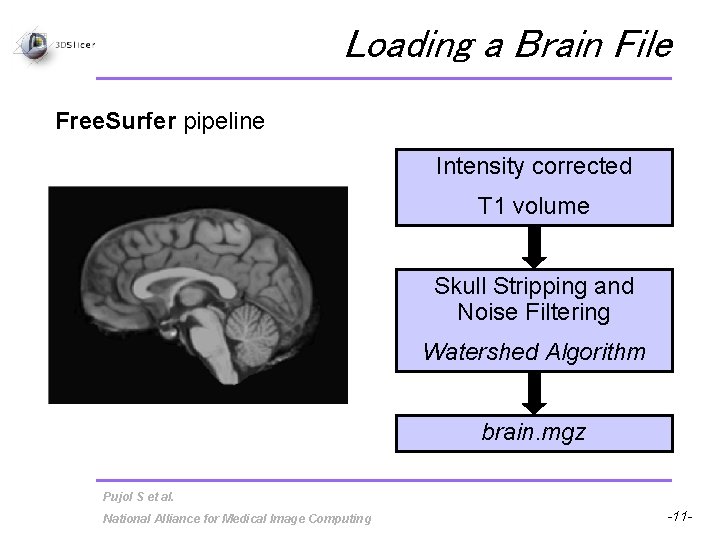
Loading a Brain File Free. Surfer pipeline Intensity corrected T 1 volume Skull Stripping and Noise Filtering Watershed Algorithm brain. mgz Pujol S et al. National Alliance for Medical Image Computing -11 -

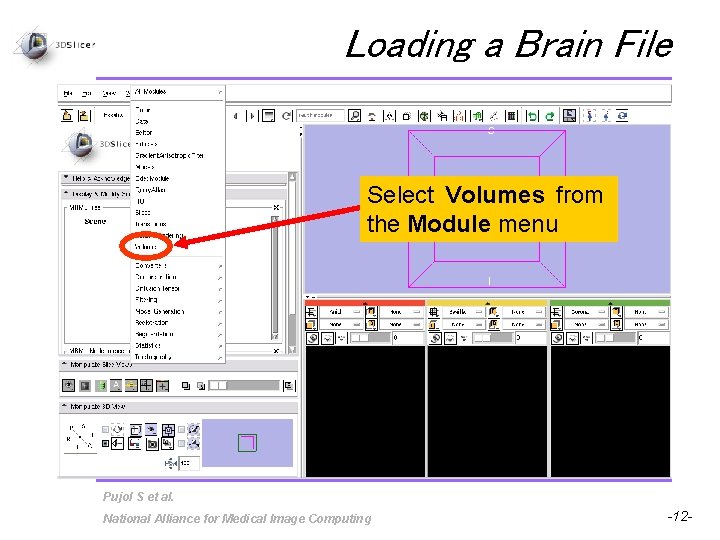
Loading a Brain File Select Volumes from the Module menu Pujol S et al. National Alliance for Medical Image Computing -12 -

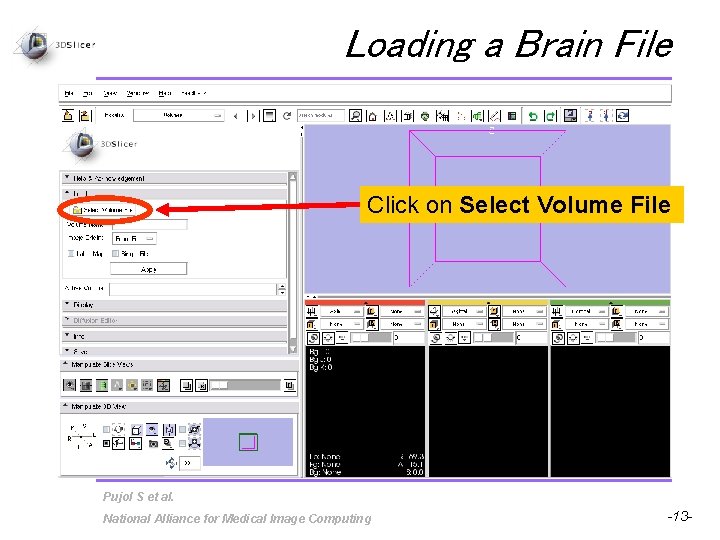
Loading a Brain File Click on Select Volume File Pujol S et al. National Alliance for Medical Image Computing -13 -

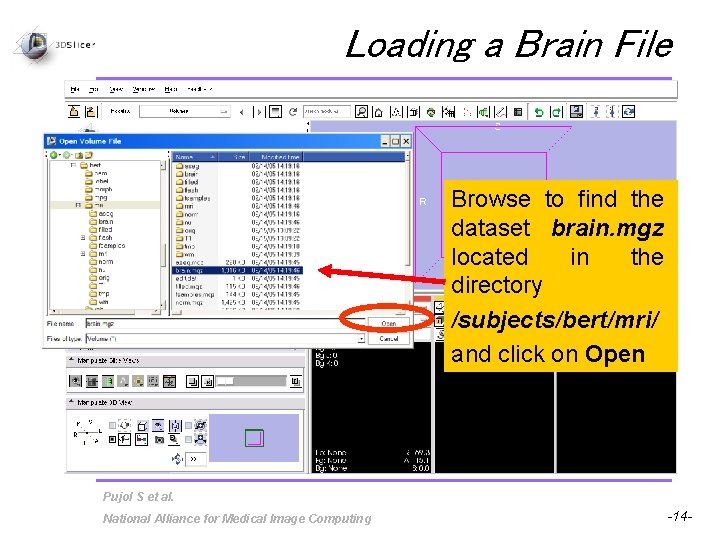
Loading a Brain File Browse to find the dataset brain. mgz located in the directory /subjects/bert/mri/ and click on Open Pujol S et al. National Alliance for Medical Image Computing -14 -

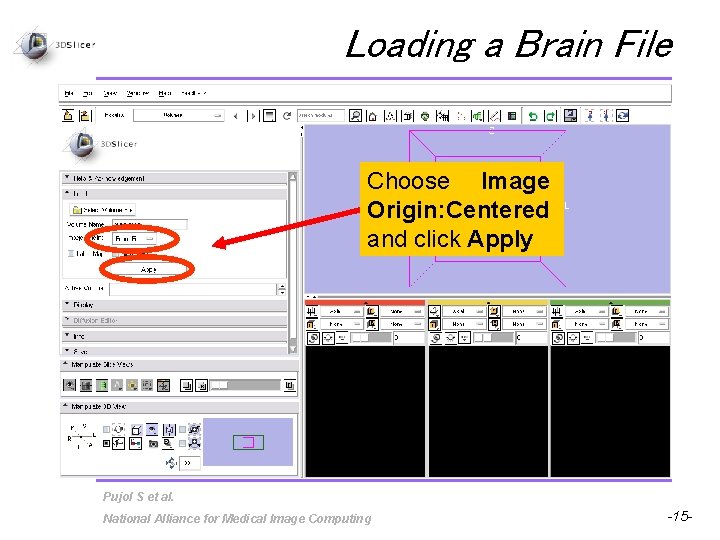
Loading a Brain File Choose Image Origin: Centered and click Apply Pujol S et al. National Alliance for Medical Image Computing -15 -

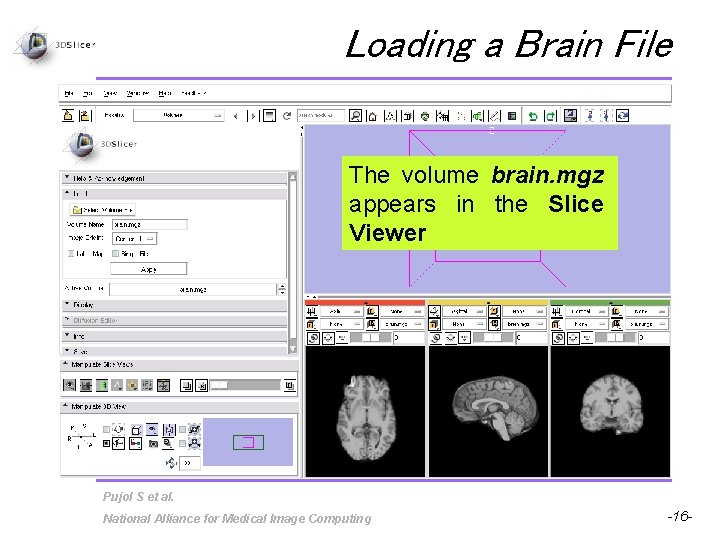
Loading a Brain File The volume brain. mgz appears in the Slice Viewer Pujol S et al. National Alliance for Medical Image Computing -16 -

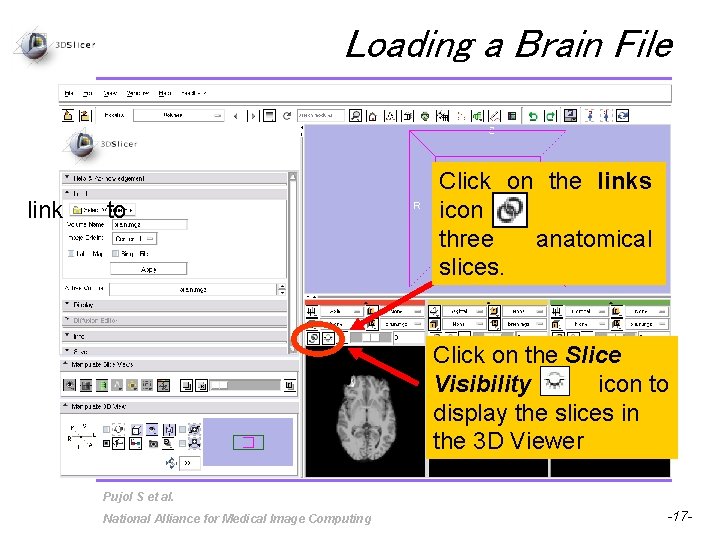
Loading a Brain File link to Click on the links icon three anatomical slices. Click on the Slice Visibility icon to display the slices in the 3 D Viewer Pujol S et al. National Alliance for Medical Image Computing -17 -

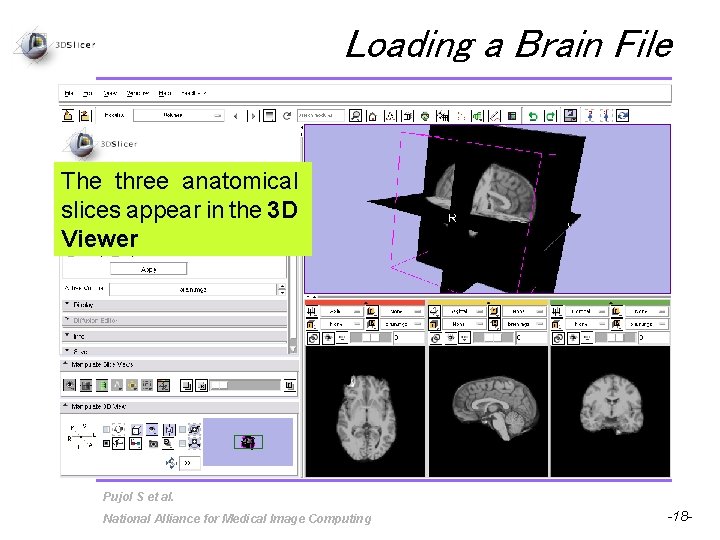
Loading a Brain File The three anatomical slices appear in the 3 D Viewer Pujol S et al. National Alliance for Medical Image Computing -18 -

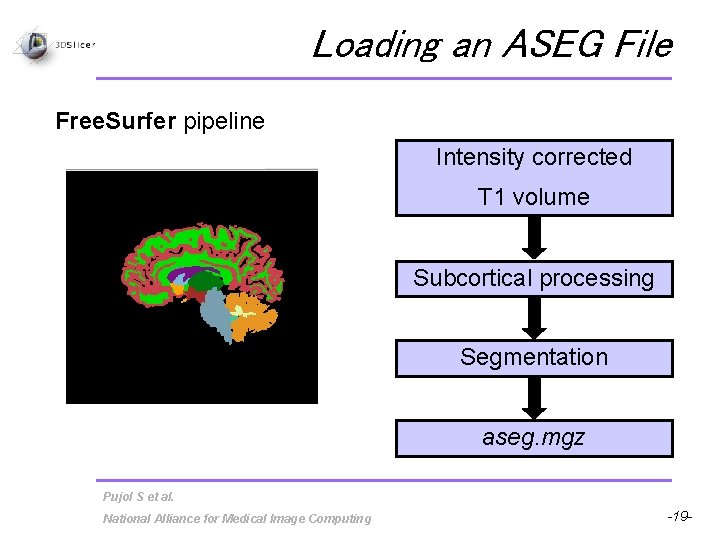
Loading an ASEG File Free. Surfer pipeline Intensity corrected T 1 volume Subcortical processing Segmentation aseg. mgz Pujol S et al. National Alliance for Medical Image Computing -19 -

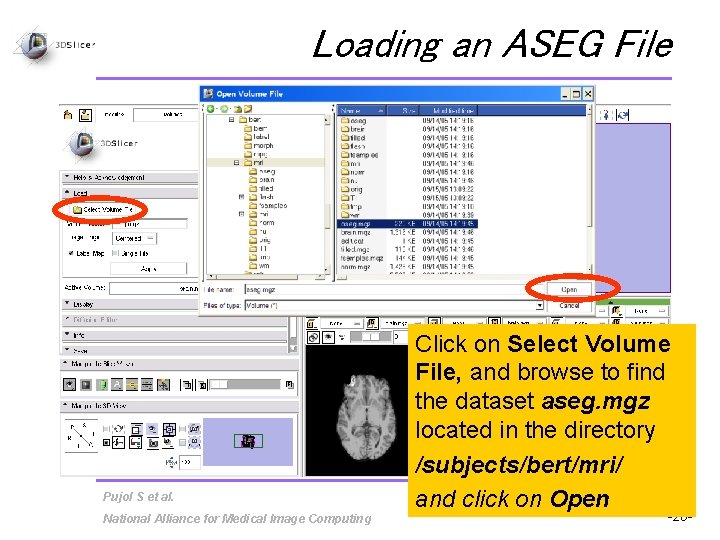
Loading an ASEG File Pujol S et al. National Alliance for Medical Image Computing Click on Select Volume File, and browse to find the dataset aseg. mgz located in the directory /subjects/bert/mri/ and click on Open -20 -

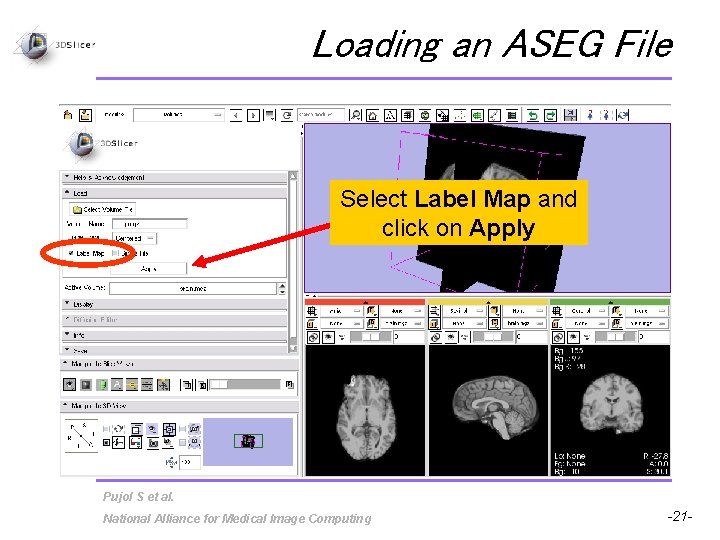
Loading an ASEG File Select Label Map and click on Apply Pujol S et al. National Alliance for Medical Image Computing -21 -

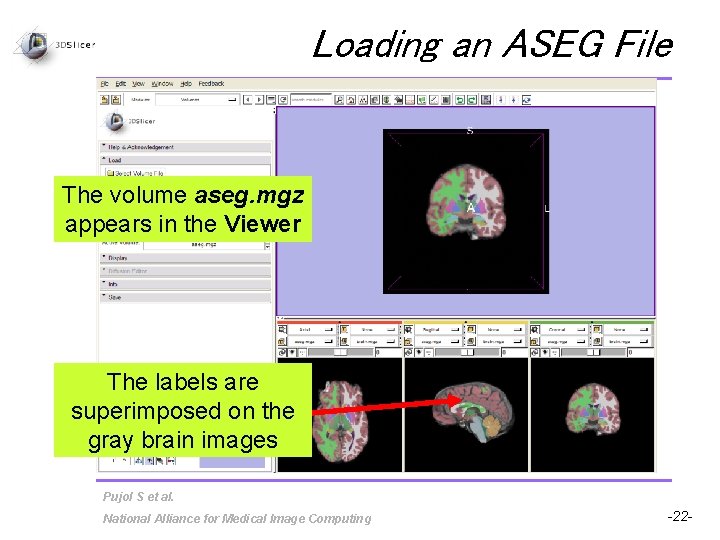
Loading an ASEG File The volume aseg. mgz appears in the Viewer The labels are superimposed on the gray brain images Pujol S et al. National Alliance for Medical Image Computing -22 -

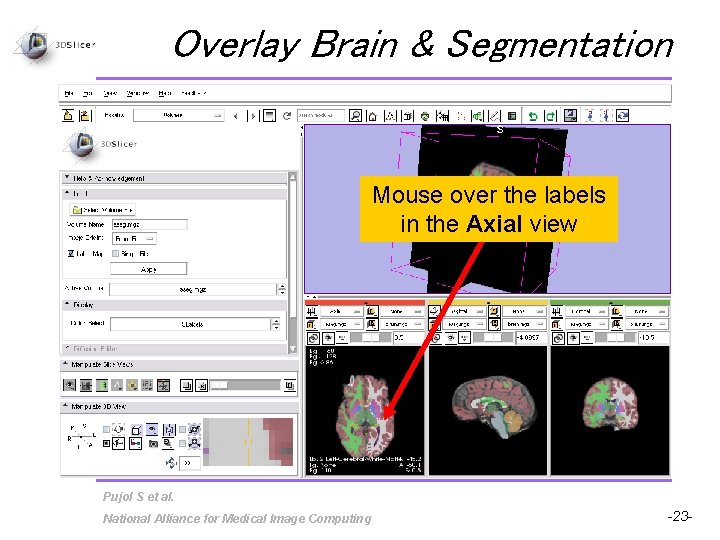
Overlay Brain & Segmentation Mouse over the labels in the Axial view Pujol S et al. National Alliance for Medical Image Computing -23 -

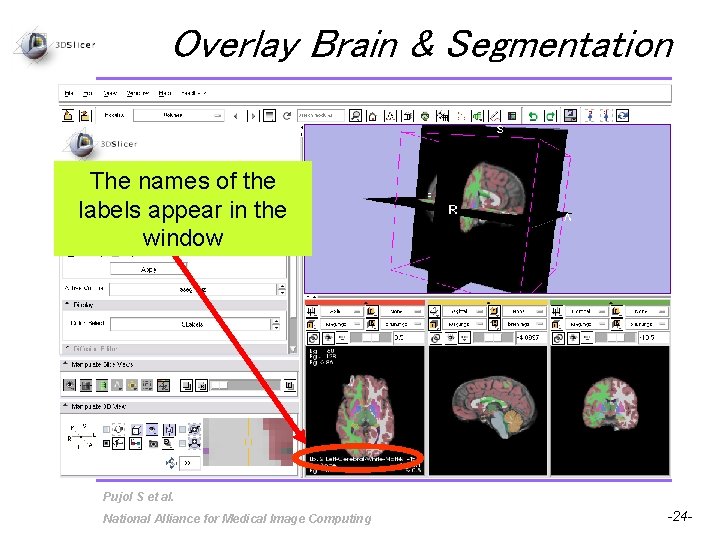
Overlay Brain & Segmentation The names of the labels appear in the window Pujol S et al. National Alliance for Medical Image Computing -24 -

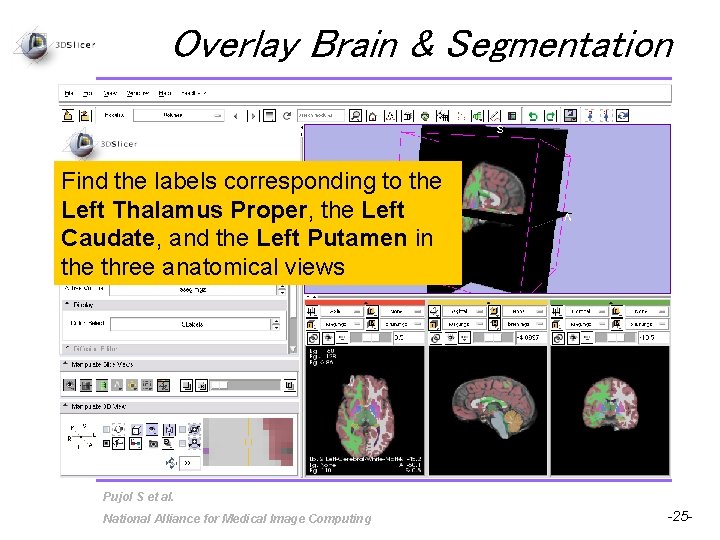
Overlay Brain & Segmentation Find the labels corresponding to the Left Thalamus Proper, the Left Caudate, and the Left Putamen in the three anatomical views Pujol S et al. National Alliance for Medical Image Computing -25 -

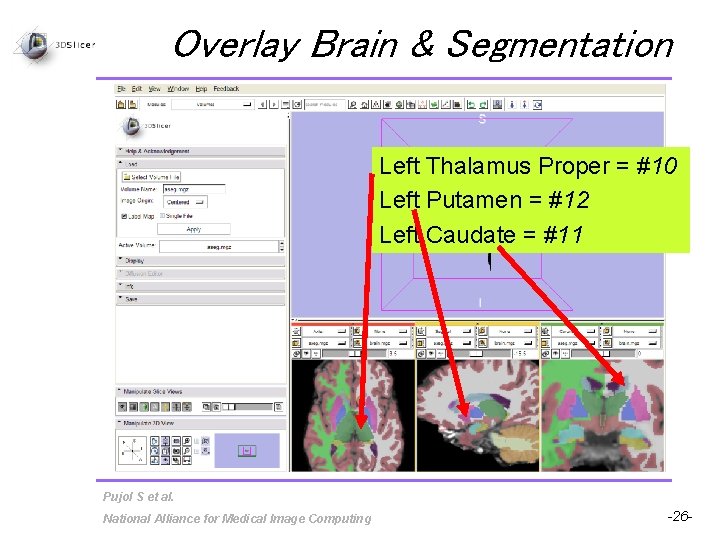
Overlay Brain & Segmentation Left Thalamus Proper = #10 Left Putamen = #12 Left Caudate = #11 Pujol S et al. National Alliance for Medical Image Computing -26 -

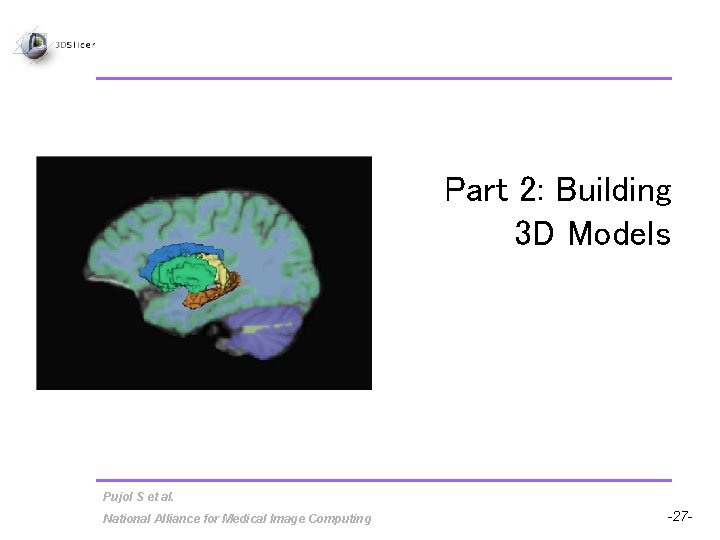
Part 2: Building 3 D Models Pujol S et al. National Alliance for Medical Image Computing -27 -

Building 3 D Models • Building a Single Model • Building Multiple Models Pujol S et al. National Alliance for Medical Image Computing -28 -

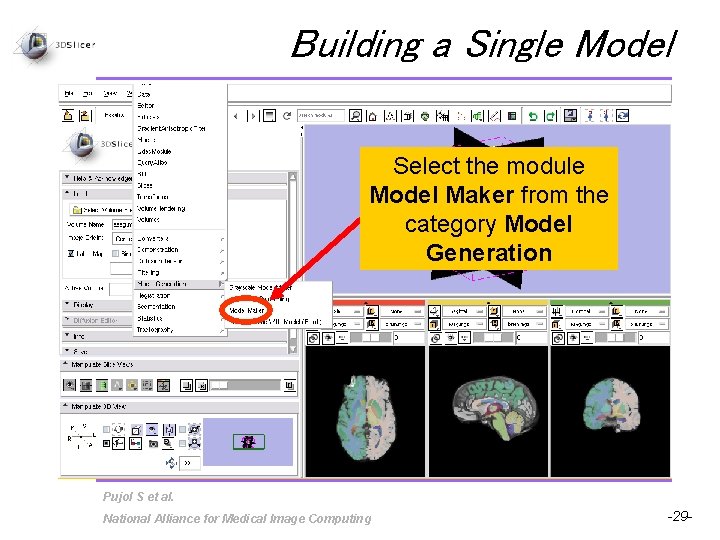
Building a Single Model Select the module Model Maker from the category Model Generation Pujol S et al. National Alliance for Medical Image Computing -29 -

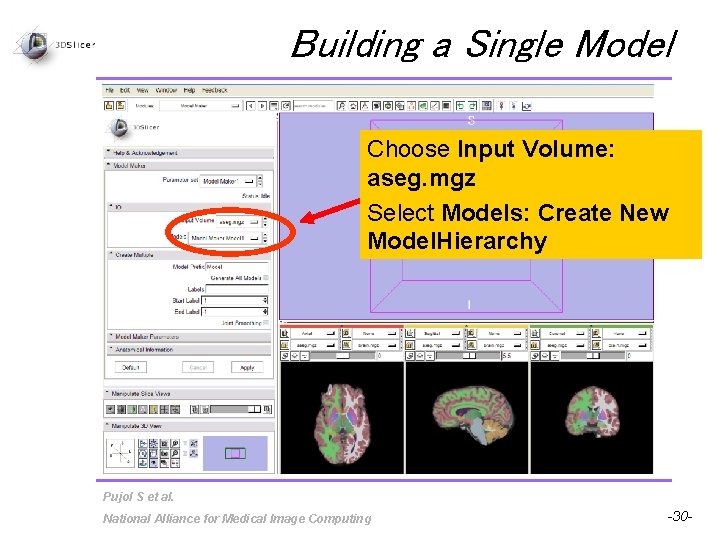
Building a Single Model Choose Input Volume: aseg. mgz Select Models: Create New Model. Hierarchy Pujol S et al. National Alliance for Medical Image Computing -30 -

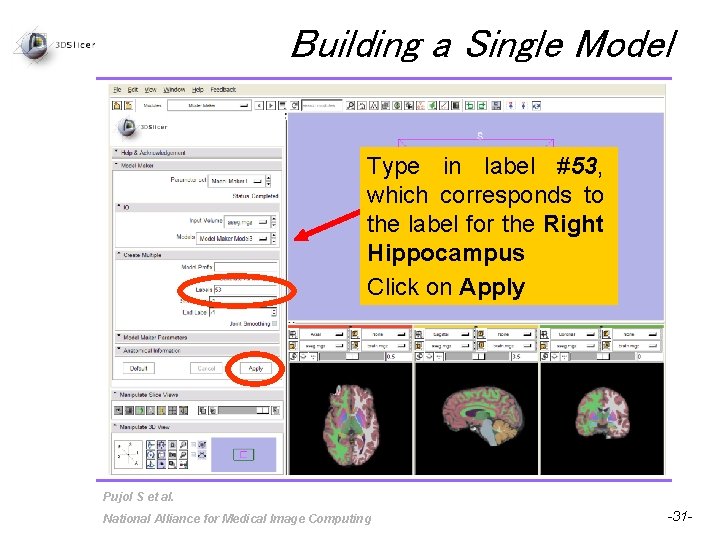
Building a Single Model Type in label #53, which corresponds to the label for the Right Hippocampus Click on Apply Pujol S et al. National Alliance for Medical Image Computing -31 -

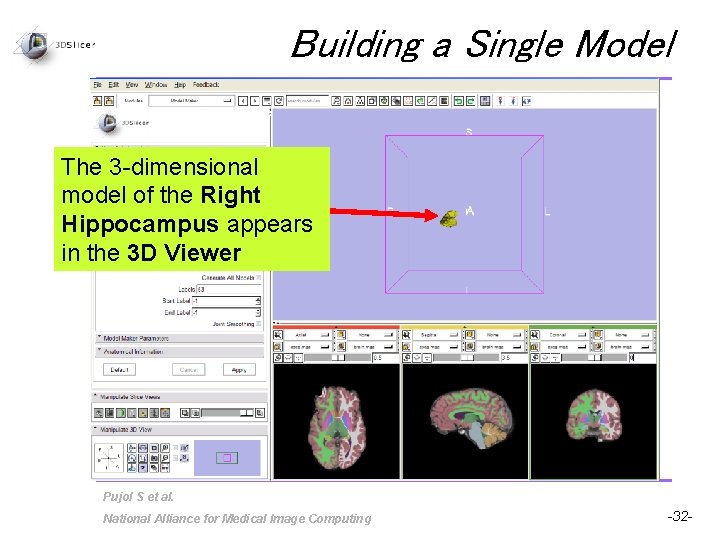
Building a Single Model The 3 -dimensional model of the Right Hippocampus appears in the 3 D Viewer Pujol S et al. National Alliance for Medical Image Computing -32 -

Building 3 D Models • Building a Single Model • Building Multiple Models Pujol S et al. National Alliance for Medical Image Computing -33 -

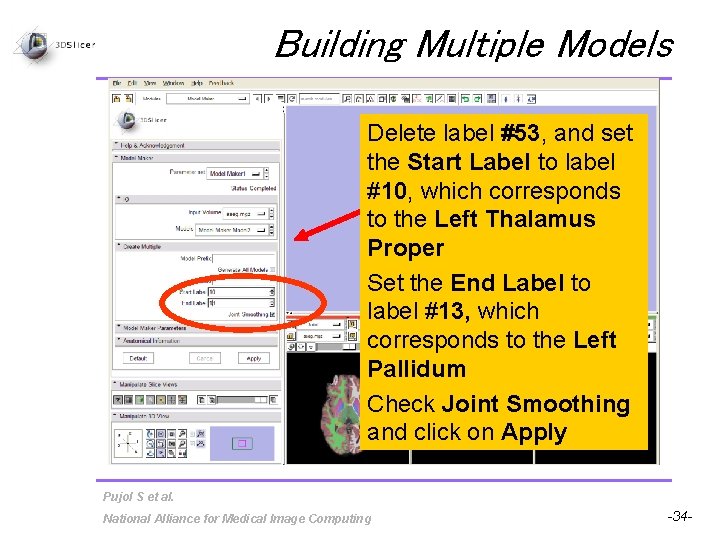
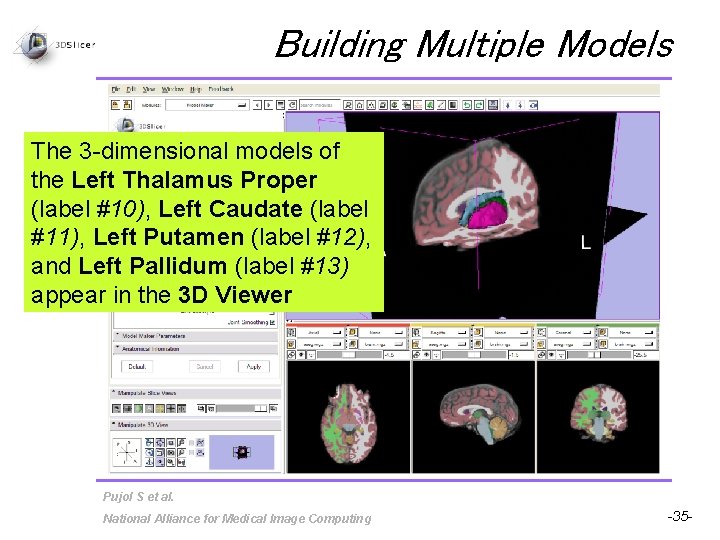
Building Multiple Models Delete label #53, and set the Start Label to label #10, which corresponds to the Left Thalamus Proper Set the End Label to label #13, which corresponds to the Left Pallidum Check Joint Smoothing and click on Apply Pujol S et al. National Alliance for Medical Image Computing -34 -

Building Multiple Models The 3 -dimensional models of the Left Thalamus Proper (label #10), Left Caudate (label #11), Left Putamen (label #12), and Left Pallidum (label #13) appear in the 3 D Viewer Pujol S et al. National Alliance for Medical Image Computing -35 -

Part 3: Loading Free. Surfer Surfaces and Visualizing Parcellation Maps Pujol S et al. National Alliance for Medical Image Computing -36 -

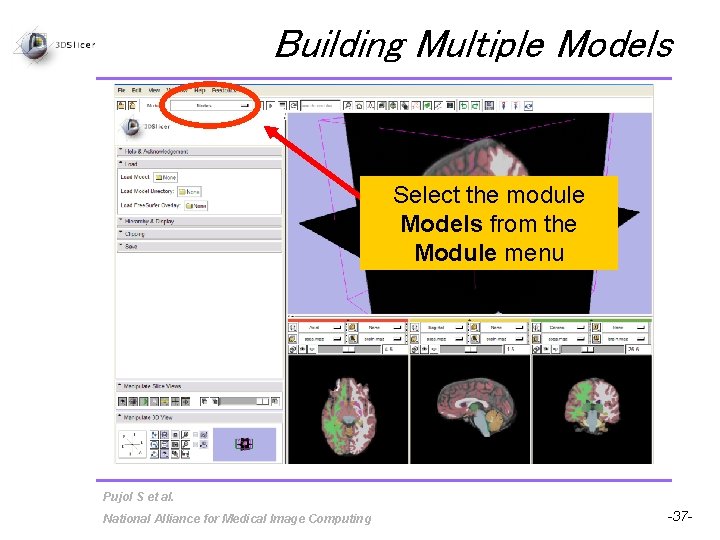
Building Multiple Models Select the module Models from the Module menu Pujol S et al. National Alliance for Medical Image Computing -37 -

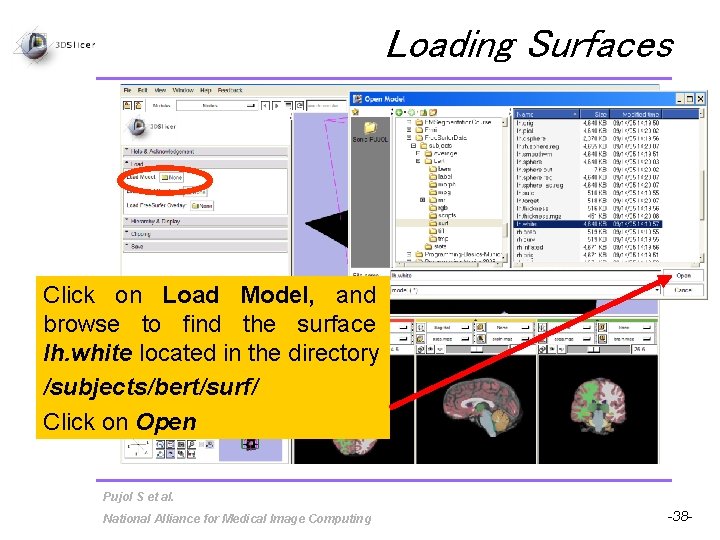
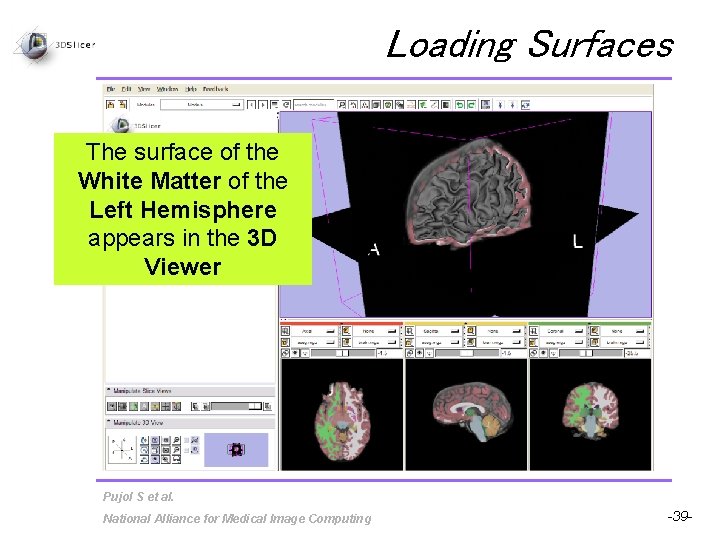
Loading Surfaces Click on Load Model, and browse to find the surface lh. white located in the directory /subjects/bert/surf/ Click on Open Pujol S et al. National Alliance for Medical Image Computing -38 -

Loading Surfaces The surface of the White Matter of the Left Hemisphere appears in the 3 D Viewer Pujol S et al. National Alliance for Medical Image Computing -39 -

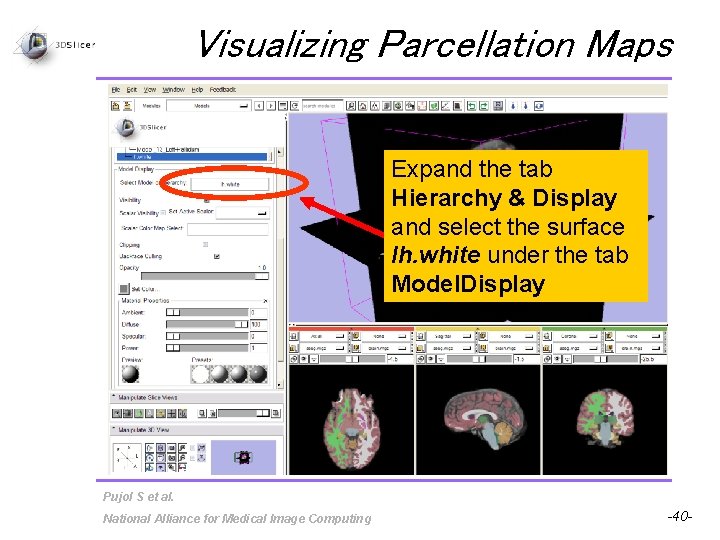
Visualizing Parcellation Maps Expand the tab Hierarchy & Display and select the surface lh. white under the tab Model. Display Pujol S et al. National Alliance for Medical Image Computing -40 -

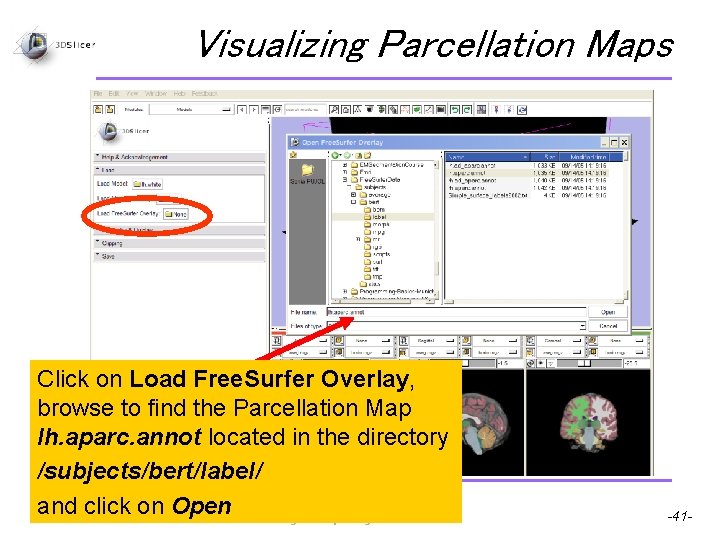
Visualizing Parcellation Maps Click on Load Free. Surfer Overlay, browse to find the Parcellation Map lh. aparc. annot located in the directory /subjects/bert/label/ Pujol S et al. and click on Open National Alliance for Medical Image Computing -41 -

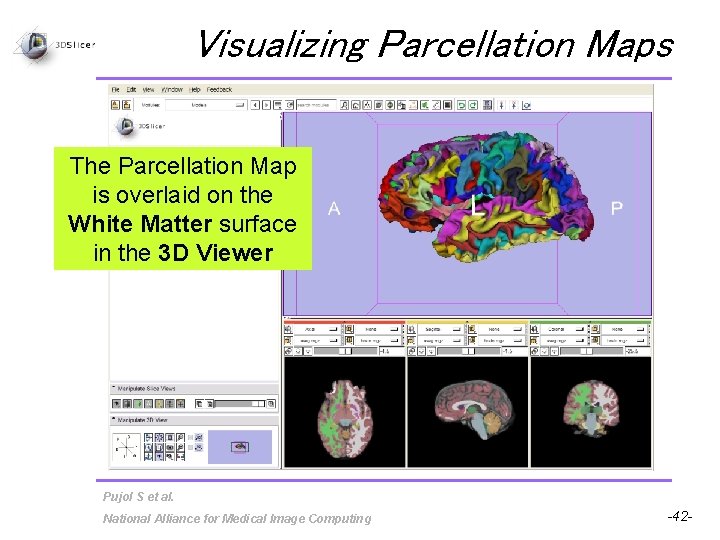
Visualizing Parcellation Maps The Parcellation Map is overlaid on the White Matter surface in the 3 D Viewer Pujol S et al. National Alliance for Medical Image Computing -42 -

Part 4: Automatic Data Loading via a Generic Scene File Pujol S et al. National Alliance for Medical Image Computing -43 -

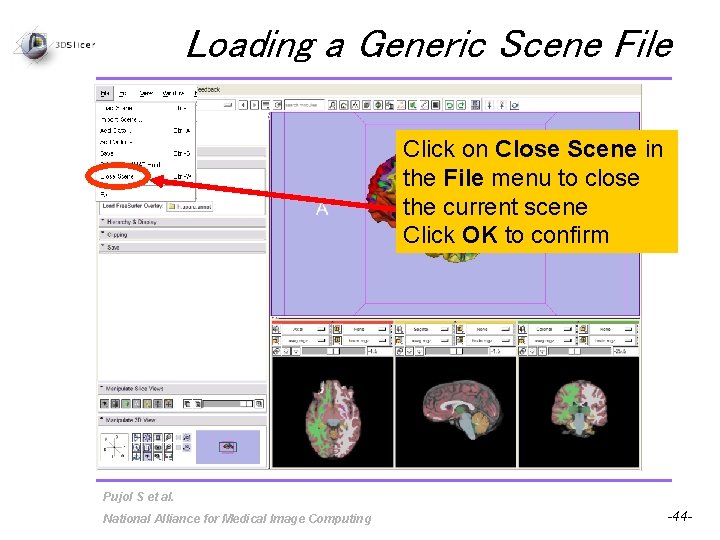
Loading a Generic Scene File Click on Close Scene in the File menu to close the current scene Click OK to confirm Pujol S et al. National Alliance for Medical Image Computing -44 -

Loading a Generic Scene File • The generic scene file works by looking in the subject directory created by Free. Surfer, and loading all available volumes and models based on known subdirectory names and filenames. • The file slicer. Generic. Scene. mrml will work properly if the subdirectory names and filenames have not been changed by the user. Pujol S et al. National Alliance for Medical Image Computing -45 -

Loading a Generic Scene File Copy the file slicer. Generic. Scene. mrml into the directory /subjects/ of our tutorial dataset. /subjects/ Pujol S et al. National Alliance for Medical Image Computing -46 -

Loading a Generic Scene File Copy the file slicer. Generic. Scene. mrml located in the directory /subjects/, into the directory /subjects/bert/ of our sample subject. /subjects/bert/ Pujol S et al. National Alliance for Medical Image Computing -47 -

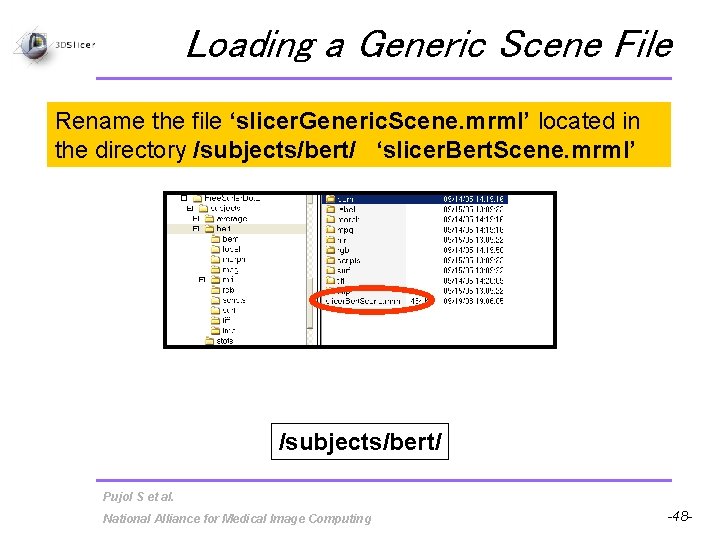
Loading a Generic Scene File Rename the file ‘slicer. Generic. Scene. mrml’ located in the directory /subjects/bert/ ‘slicer. Bert. Scene. mrml’ /subjects/bert/ Pujol S et al. National Alliance for Medical Image Computing -48 -

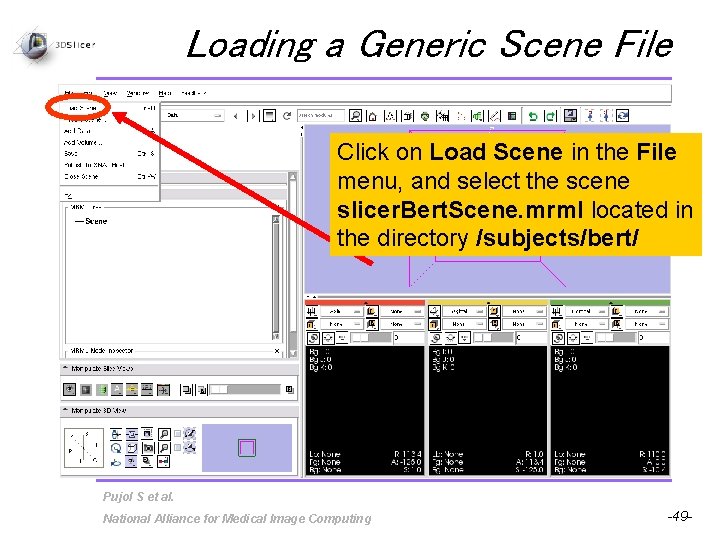
Loading a Generic Scene File Click on Load Scene in the File menu, and select the scene slicer. Bert. Scene. mrml located in the directory /subjects/bert/ Pujol S et al. National Alliance for Medical Image Computing -49 -

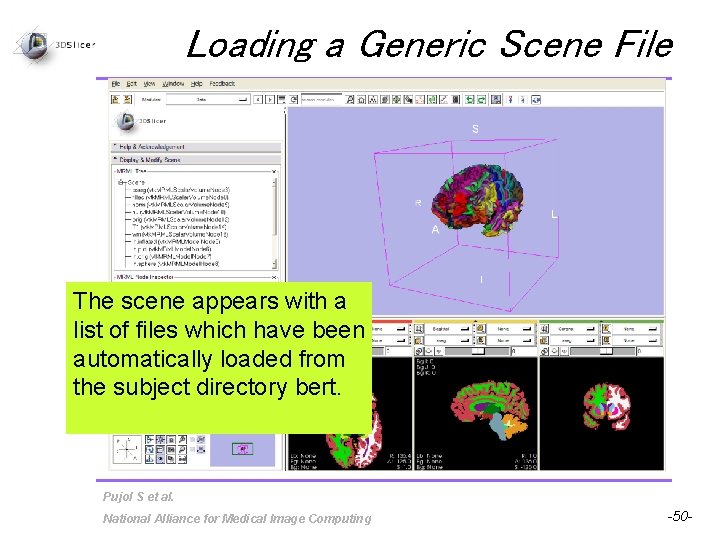
Loading a Generic Scene File The scene appears with a list of files which have been automatically loaded from the subject directory bert. Pujol S et al. National Alliance for Medical Image Computing -50 -

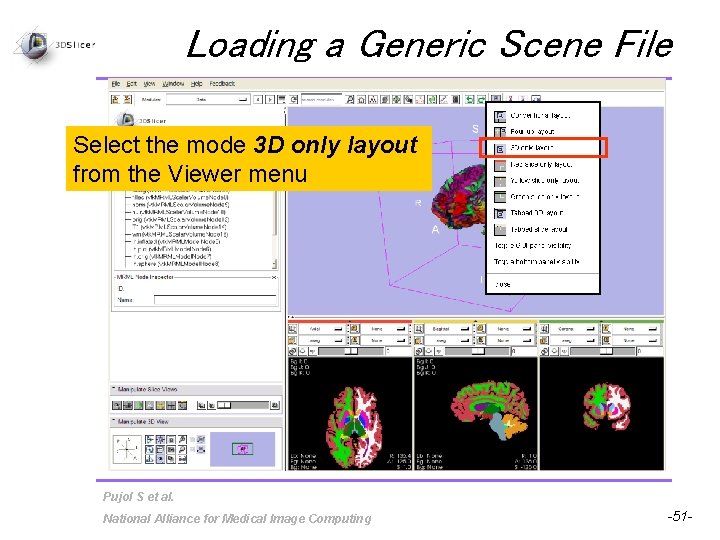
Loading a Generic Scene File Select the mode 3 D only layout from the Viewer menu Pujol S et al. National Alliance for Medical Image Computing -51 -

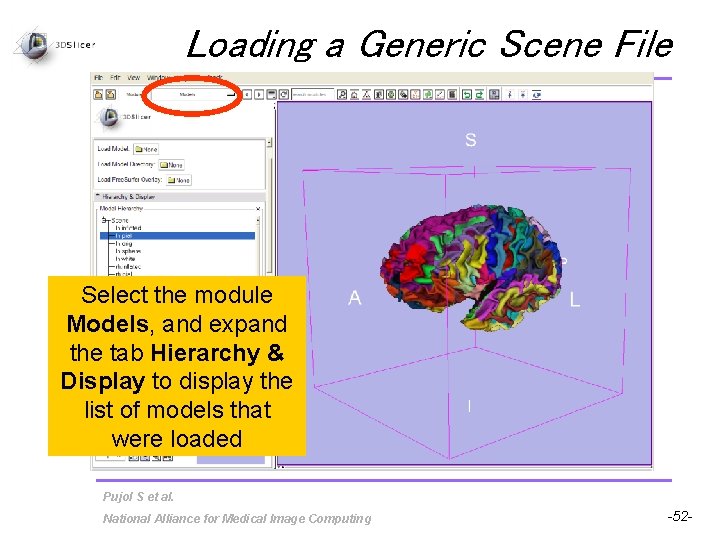
Loading a Generic Scene File Select the module Models, and expand the tab Hierarchy & Display to display the list of models that were loaded Pujol S et al. National Alliance for Medical Image Computing -52 -

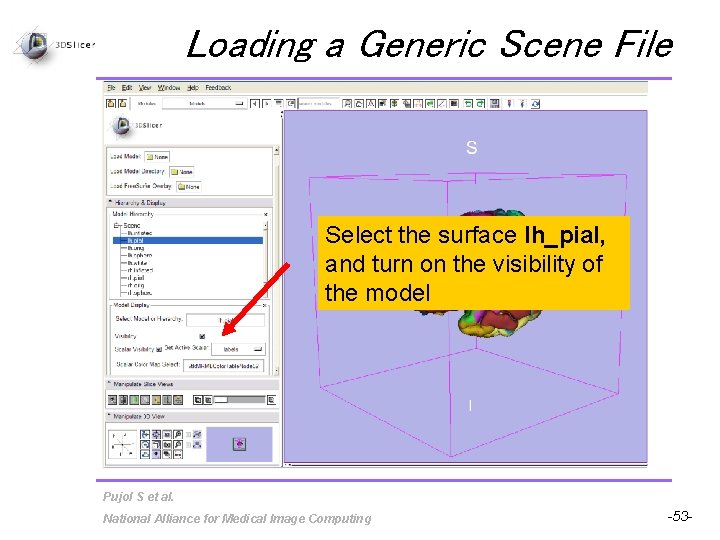
Loading a Generic Scene File Select the surface lh_pial, and turn on the visibility of the model Pujol S et al. National Alliance for Medical Image Computing -53 -

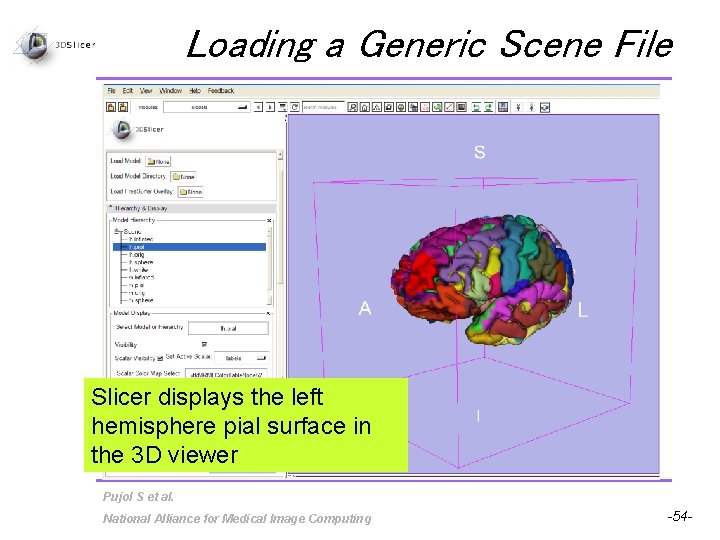
Loading a Generic Scene File Slicer displays the left hemisphere pial surface in the 3 D viewer Pujol S et al. National Alliance for Medical Image Computing -54 -

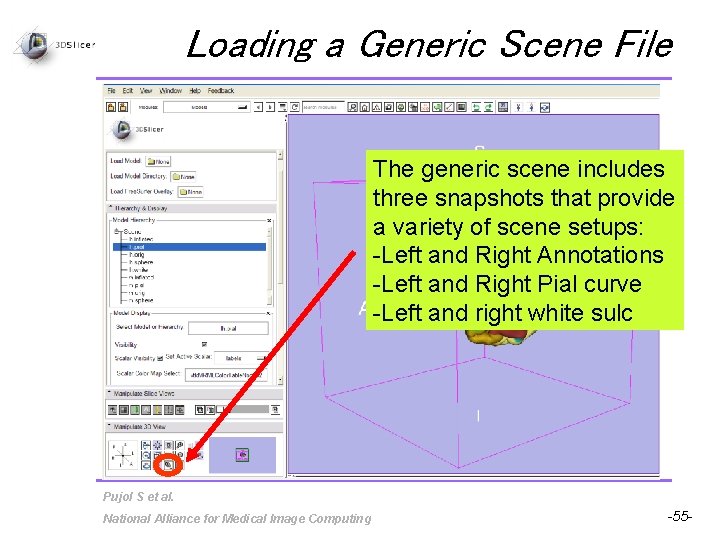
Loading a Generic Scene File The generic scene includes three snapshots that provide a variety of scene setups: -Left and Right Annotations -Left and Right Pial curve -Left and right white sulc Pujol S et al. National Alliance for Medical Image Computing -55 -

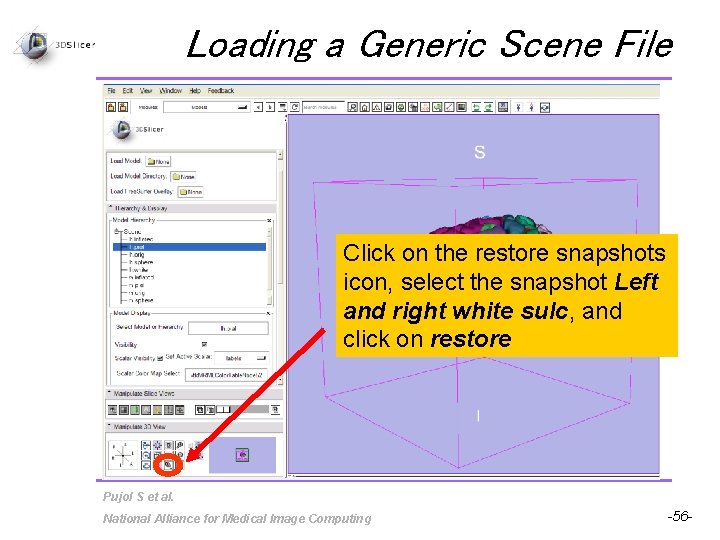
Loading a Generic Scene File Click on the restore snapshots icon, select the snapshot Left and right white sulc, and click on restore Pujol S et al. National Alliance for Medical Image Computing -56 -

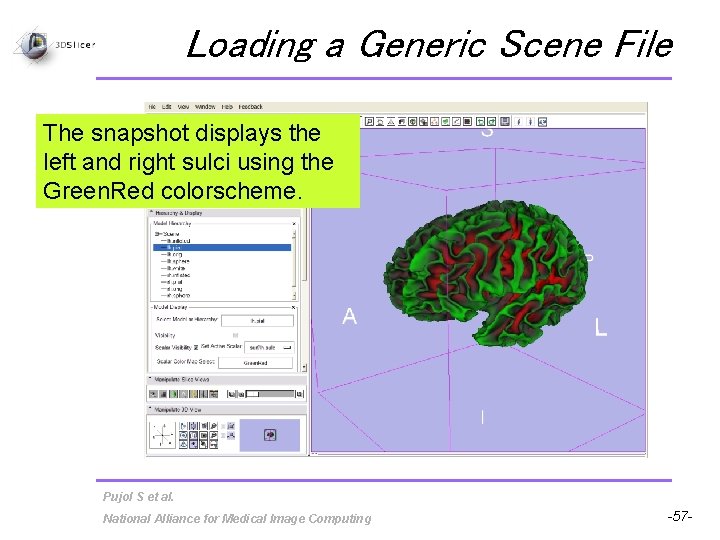
Loading a Generic Scene File The snapshot displays the left and right sulci using the Green. Red colorscheme. Pujol S et al. National Alliance for Medical Image Computing -57 -

Summary From Free. Surfer, Slicer 3 can load: • Brain volumes. . . • ASEG volumes. . . . • Surfaces. . . . • Parcellation Maps. . . . • All of the above, via a scene file. . . . Pujol S et al. National Alliance for Medical Image Computing -58 -

Conclusion • 3 D visualization of brain segmented surfaces and parcellation maps • Intuitive graphical user interface to interact with Free. Surfer data • Multi platforms open-source environment spujol@bwh. harvard. edu Pujol S et al. National Alliance for Medical Image Computing -59 -