2018 MFMER slide1 2018 MFMER slide2 3 2018








- Slides: 12

© 2018 MFMER | slide-1

© 2018 MFMER | slide-2

3 © 2018 MFMER | slide-3

4 © 2018 MFMER | slide-4

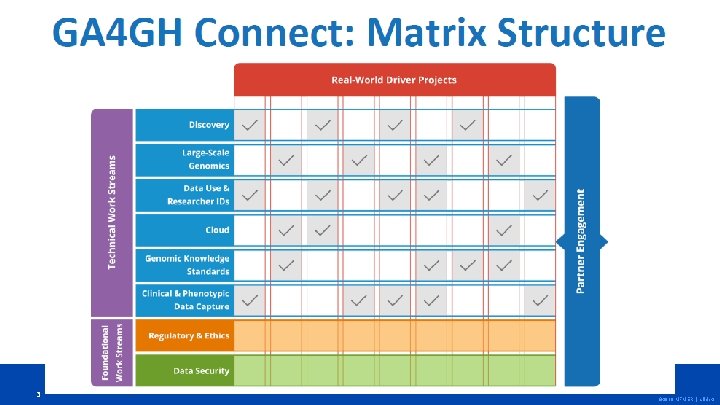
Genomic Knowledge Standards GKS aims to develop, adopt, and adapt standards to enable the exchange of genomic knowledge • • 5 Establish framework of standards that lower barriers to translating genomic knowledge into clinical practice Reusing existing knowledge is essential to future impact Knowledge repositories are a key component Collaborate with other SDOs on harmonized specifications © 2018 MFMER | slide-5

Genomic Knowledge Standards Variant Annotation • DP priorities => WS goal • • 6 • Represent and link annotations, including their evidence and provenance, to variants Supports clinical (lab) interpretation Structured, curated data from publications Clinical relevance, disease association Draft spec by Q 4 2018 © 2018 MFMER | slide-6

Genomic Knowledge Standards Variant Representation • DP priorities => WS goal • • • Simple variants (VMC) • • v 1. 0 by Q 4 2018 Peer-reviewed publication Complex variants (structural, CNV, imprecise ends) • 7 Create a standard model for computer readable variant representation (definitional) v 0. 1 by Q 4 2018 © 2018 MFMER | slide-7

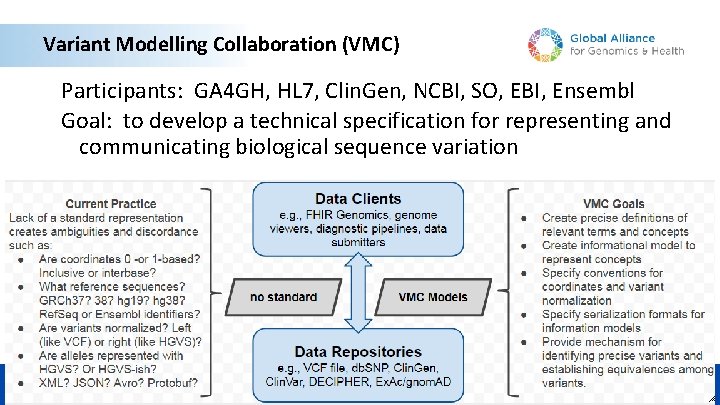
Variant Modelling Collaboration (VMC) Participants: GA 4 GH, HL 7, Clin. Gen, NCBI, SO, EBI, Ensembl Goal: to develop a technical specification for representing and communicating biological sequence variation © 2018 MFMER | slide-8

VMC 1. 0 Focus: variation that is defined by alleles — assertions of a specific sequence at precise locations on a sequence Entities: Sequence, Location, Allele, Haplotype, Genotype • • • Terminology Information Model – Minimalistic, use as part of a given use case Machine-readable schema definition © 2018 MFMER | slide-9

Sync for Genes • Sync for Genes (S 4 G) is collaboration between ONC, NIH, and industry to help deliver on goals of the Precision Medicine Initiative (PMI) and NIH’s All of Us Research Program. • Goal to standardize the sharing of genomic information between labs, providers, patients and researchers • Develop and build upon Health Level Seven International (HL 7)’s Fast Healthcare Interoperability Resources (FHIR) Genomic specifications 10

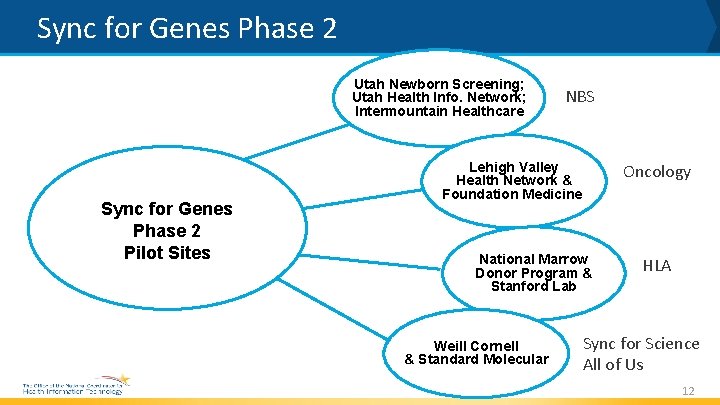
Sync for Genes Phase 2 • Takes the next step from use case and standard development (Phase 1) to actual implementation and use » Phase 1 final report: https: //www. healthit. gov/sites/default/files/sync_for_genes_report_november _2017. pdf • Pilots selected to participate in Phase 2 will focus on: » expanding the FHIR genomics profile work developed under phase 1; » addressing additional core use cases from the DAM; and » demonstrating the integration of genomics information into a clinical setting for supporting care and research efforts 11

Sync for Genes Phase 2 Utah Newborn Screening; Utah Health Info. Network; Intermountain Healthcare Sync for Genes Phase 2 Pilot Sites NBS Lehigh Valley Health Network & Foundation Medicine Oncology National Marrow Donor Program & Stanford Lab Weill Cornell & Standard Molecular HLA Sync for Science All of Us 12