2012 2 11 Chapter 8 SDSPAGE Analysis and





![example [Fig. 1] Rf = migration distance of the protein/ migration distance of the example [Fig. 1] Rf = migration distance of the protein/ migration distance of the](https://slidetodoc.com/presentation_image_h/aacaee8ce488915179f0465928307e2e/image-20.jpg)




- Slides: 27

2012 - 생화학 실험 (2) 11주차 Chapter 8. SDS-PAGE Analysis and Molecular Weight Determination 담당교수 : 하상준 교수님(S 423) 담당조교 : 배우리(S 329), 임서진 (S 338)

Purpose • Understanding the principles of polyacrylamide gel electrophoresis (PAGE) • Construct standard curves and determine protein concentrations by the Bradford method

I. PAGE

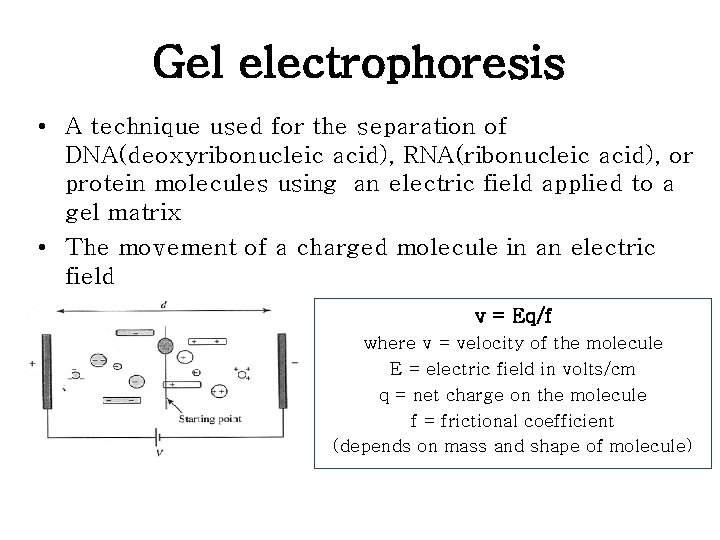
Gel electrophoresis • A technique used for the separation of DNA(deoxyribonucleic acid), RNA(ribonucleic acid), or protein molecules using an electric field applied to a gel matrix • The movement of a charged molecule in an electric field v = Eq/f where v = velocity of the molecule E = electric field in volts/cm q = net charge on the molecule f = frictional coefficient (depends on mass and shape of molecule)

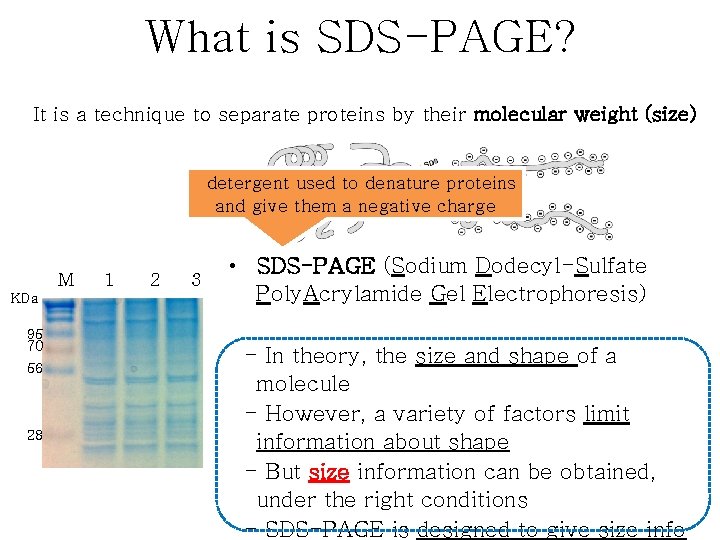
What is SDS-PAGE? It is a technique to separate proteins by their molecular weight (size) detergent used to denature proteins and give them a negative charge M KDa 95 70 56 28 1 2 3 • SDS-PAGE (Sodium Dodecyl-Sulfate Poly. Acrylamide Gel Electrophoresis) - In theory, the size and shape of a molecule - However, a variety of factors limit information about shape - But size information can be obtained, under the right conditions - SDS-PAGE is designed to give size info

Polyacrylamide Gels (PAG) • • Gel of choice for protein separations Prepared by the free radical polymerization of acrylamide and N, N’-methylene-bis-acrylamide Polymerization is controlled by an initiatorcatalyst system, ammonium persulfate (APS) and TEtra. Methyl. Ethylene. Diamine (TEMED) Advantages - Good resolving power for small to medium sized proteins (best for 30, 000 to 300, 000) - Useful with large sample sizes - Minimal interactions of protein with matrix - Good physical stability of the matrix - Pore sizes can be selected by varying the concentration of cross-linker

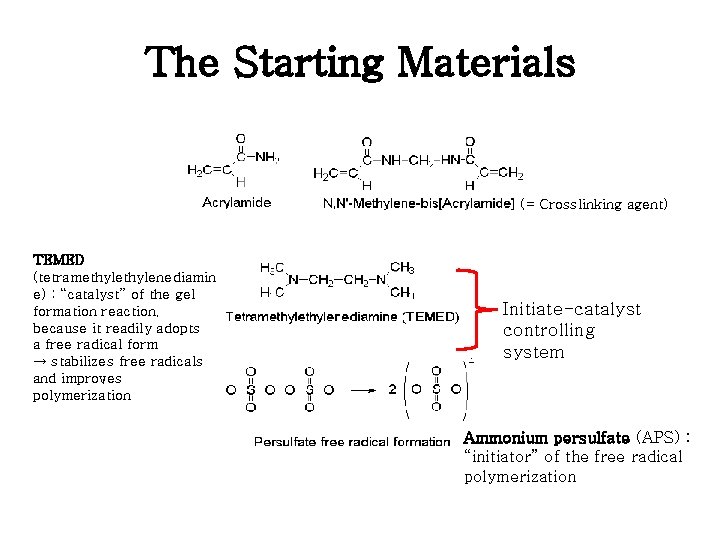
The Starting Materials (= Crosslinking agent) TEMED (tetramethylenediamin e) : “catalyst” of the gel formation reaction, because it readily adopts a free radical form → stabilizes free radicals and improves polymerization Initiate-catalyst controlling system Ammonium persulfate (APS) : “initiator” of the free radical polymerization

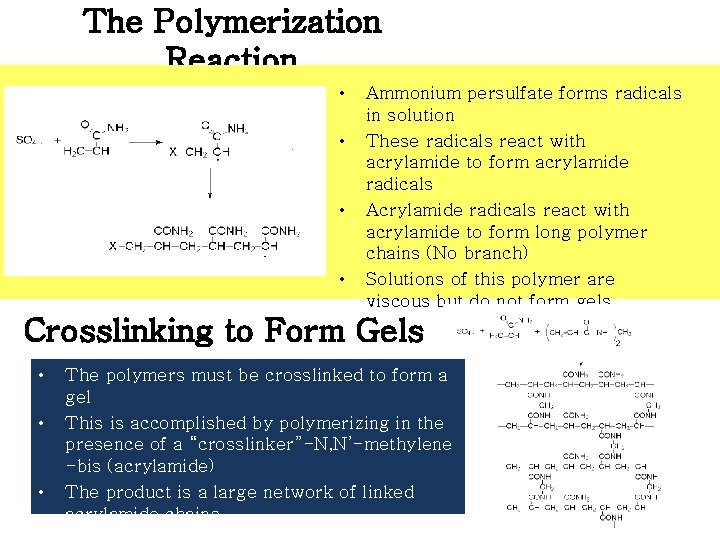
The Polymerization Reaction • • Ammonium persulfate forms radicals in solution These radicals react with acrylamide to form acrylamide radicals Acrylamide radicals react with acrylamide to form long polymer chains (No branch) Solutions of this polymer are viscous but do not form gels Crosslinking to Form Gels • • • The polymers must be crosslinked to form a gel This is accomplished by polymerizing in the presence of a “crosslinker”-N, N’-methylene -bis (acrylamide) The product is a large network of linked acrylamide chains

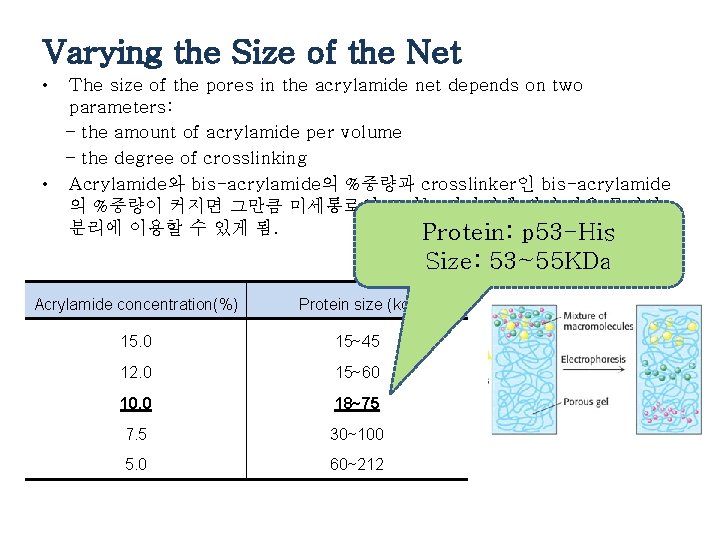
Varying the Size of the Net • The size of the pores in the acrylamide net depends on two parameters: – the amount of acrylamide per volume – the degree of crosslinking • Acrylamide와 bis-acrylamide의 %중량과 crosslinker인 bis-acrylamide 의 %중량이 커지면 그만큼 미세통로의 크기는 작아지게 되어 작은 물질의 분리에 이용할 수 있게 됨. Protein: p 53 -His Size: 53~55 KDa Acrylamide concentration(%) Protein size (kd) 15. 0 15~45 12. 0 15~60 10. 0 18~75 7. 5 30~100 5. 0 60~212

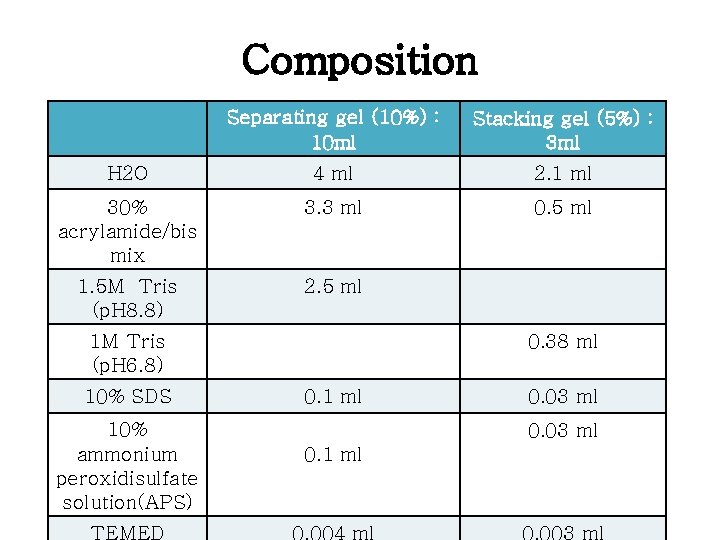
Composition Separating gel (10%) : 10 ml Stacking gel (5%) : 3 ml H 2 O 4 ml 2. 1 ml 30% acrylamide/bis mix 3. 3 ml 0. 5 ml 1. 5 M Tris (p. H 8. 8) 2. 5 ml 1 M Tris (p. H 6. 8) 10% SDS 10% ammonium peroxidisulfate solution(APS) TEMED 0. 38 ml 0. 1 ml 0. 03 ml 0. 1 ml 0. 004 ml 0. 003 ml


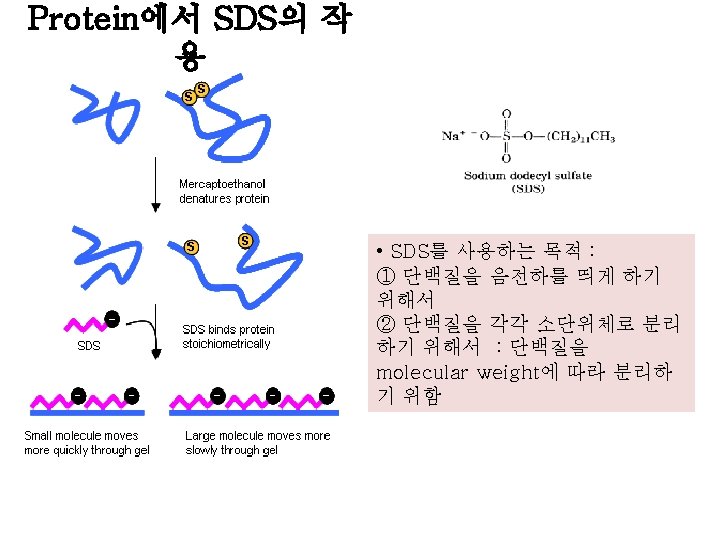
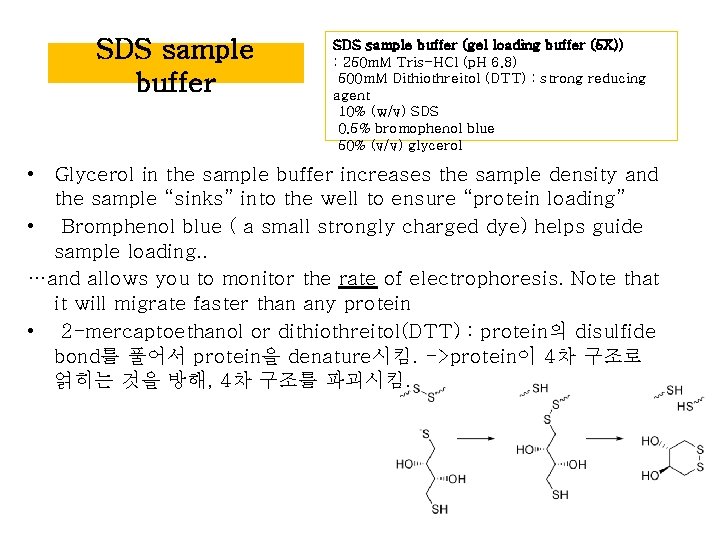
SDS sample buffer (gel loading buffer (5 X)) : 250 m. M Tris-HCl (p. H 6. 8) 500 m. M Dithiothreitol (DTT) : strong reducing agent 10% (w/v) SDS 0. 5% bromophenol blue 50% (v/v) glycerol • Glycerol in the sample buffer increases the sample density and the sample “sinks” into the well to ensure “protein loading” • Bromphenol blue ( a small strongly charged dye) helps guide sample loading. . …and allows you to monitor the rate of electrophoresis. Note that it will migrate faster than any protein • 2 -mercaptoethanol or dithiothreitol(DTT) : protein의 disulfide bond를 풀어서 protein을 denature시킴. ->protein이 4차 구조로 얽히는 것을 방해, 4차 구조를 파괴시킴.


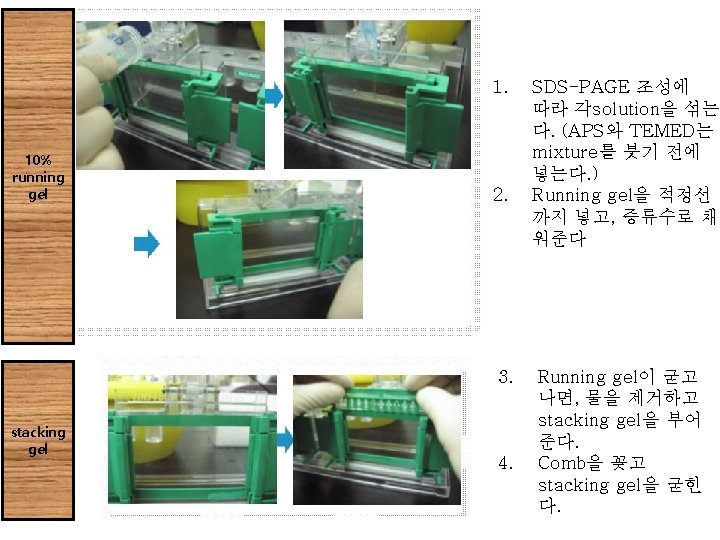
Procedure


5. Comb을 제거하고, gel을 electrophoresis kit에 casting하고 running buffer를 채워준다 6. size-marker와 sample을 20 ul loading하고, electrophoresis * Sample처리 (with 1 X SDS sample buffer) - 95℃, 10 min heat

7. Gel running이 끝나면, caster에서 gel 만 분리하여 CBB로 staining한다. Staining SDS-polyacrylamide gels with Coomassie brilliant blue(CBB) : PAGE에 의해 분리된 protein은 CBB나 silver salt로 staining하여 detection한다. CBB는 gel이 아닌 protein에 nonspecific binding을 하여 blue band로 나타난다. CBB는 aminotriarylmethanedye이며, protein 과 covalent complex가 아닌 주로van der Waals force electrostatic interaction을 NH 3+ group과 강하게 형성한다. Dye uptake는 protein 양에 비례한다. 8. Destaining solution으로 destaining하여 band만 나 타나게 한다.

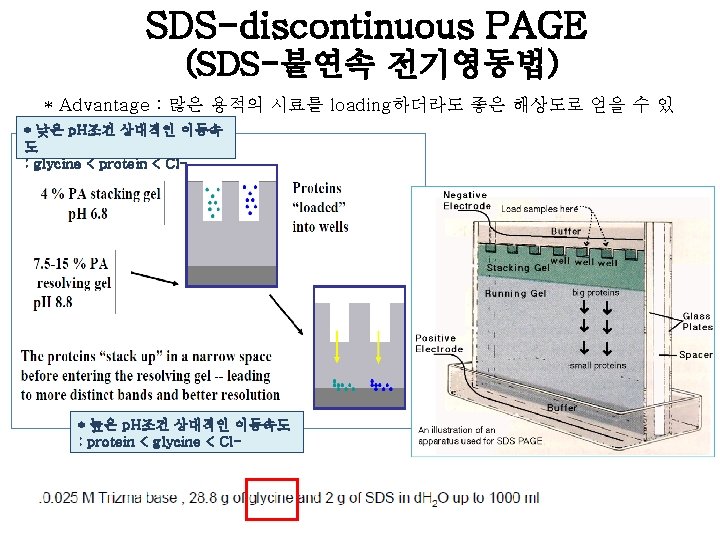
Procedure 1. SDS-PAGE 조성에 따라 각solution을 섞는다. (APS와 TEMED는mixture를 붓기 전에 넣는다. ) 2. Running gel을 적정선까지 넣고, 증류수로 채워준다. 3. Running gel이 굳고나면, 물을 제거하고 stacking gel을 부어 준다. 4. Comb을 꽂고 stacking gel을 굳힌다. 5. Comb을 제거하고, gel을 electrophoresis kit에 casting하고 running buffer를 채워준다. 6. size-marker와 sample을 loading하고, electrophoresis. 7. Gel running이 끝나면, caster에서 gel만 분리하여 CBB로 staining한다. 8. Destaining solution으로 destaining하여 band만 나타나게 한 다.

![example Fig 1 Rf migration distance of the protein migration distance of the example [Fig. 1] Rf = migration distance of the protein/ migration distance of the](https://slidetodoc.com/presentation_image_h/aacaee8ce488915179f0465928307e2e/image-20.jpg)
example [Fig. 1] Rf = migration distance of the protein/ migration distance of the dye front [Fig. 2]

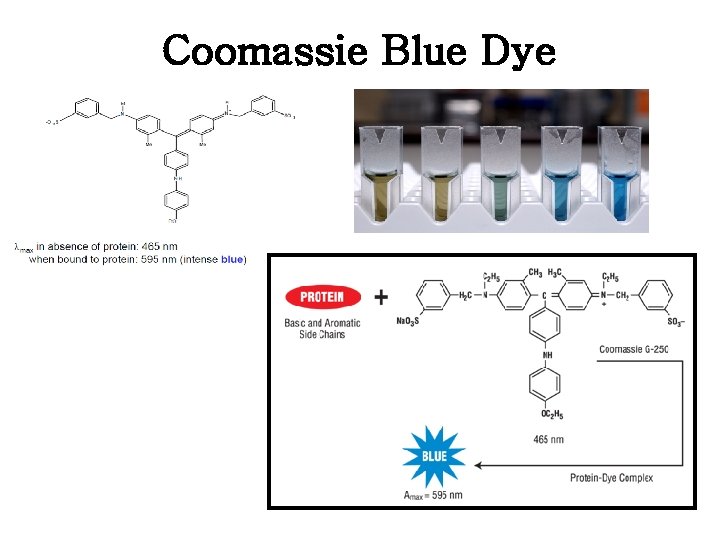
II. Bradford Assay - Coomassie Blue Dye

• Bradford assay란? - UV-visible spectrophotometer를 이용, 흡광도를 측정하여 standard물질인 BSA(bovine serum albumin)나 BGG(Bovine gamma globulin)를 기준으로 자신의 시료에 단백질이 얼마만큼 들어있는지 측정하는 방법 중 하나. 신속, 간편하게 단백질 정량 가능하여 널리 사용. Dye로는 Coomassie Blue G-250를 사용하며, 이 dye가 단백질과 결합해 생기는 파 장의 변화 측정. • A max of CB G-250 shifts from 465 to 595 nm when bound to protein - dye reacts primarily with Arg - lesser extent with His, Lys, Tyr, Trp, Phe • • sensitivity is 1 -100 mg/ml depending on circumstances single step and few interfering substances protein concentration extrapolated from standard curve sample not recoverable

Coomassie Blue Dye

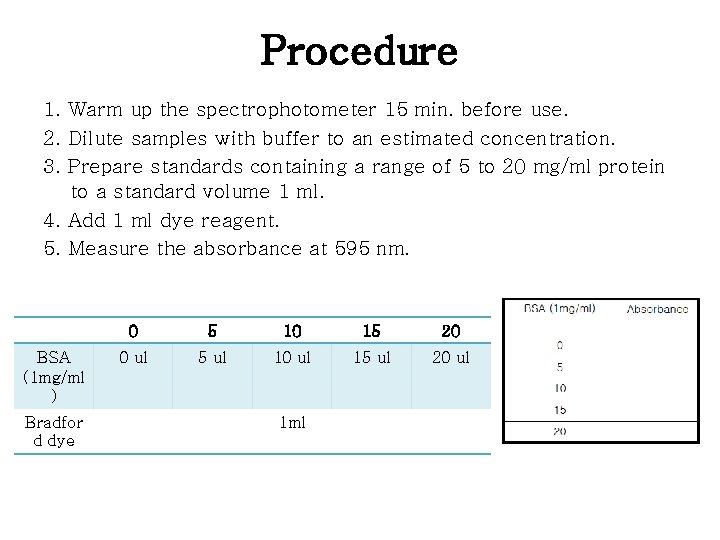
Procedure 1. Warm up the spectrophotometer 15 min. before use. 2. Dilute samples with buffer to an estimated concentration. 3. Prepare standards containing a range of 5 to 20 mg/ml protein to a standard volume 1 ml. 4. Add 1 ml dye reagent. 5. Measure the absorbance at 595 nm. BSA (1 mg/ml ) Bradfor d dye 0 5 10 15 20 0 ul 5 ul 10 ul 15 ul 20 ul 1 ml

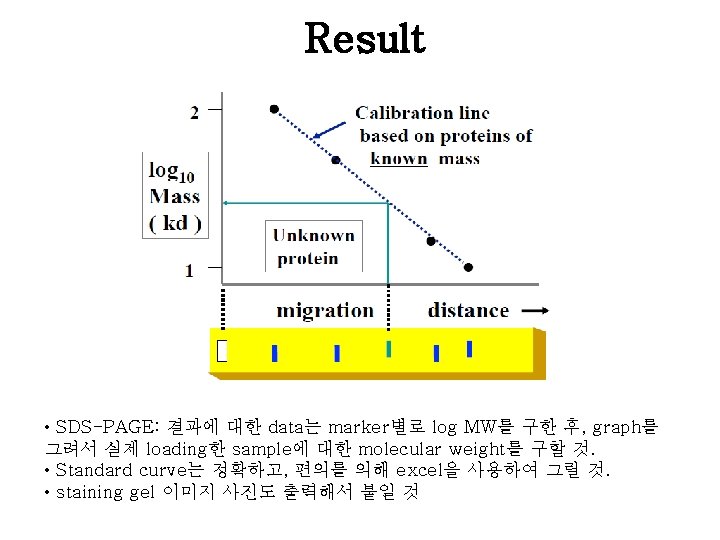
Result: Standard curve Result: Bradford Assay <Standard curve example> Samples Absorbanc e concentratio n Unknown sample #1 Unknown sample #2 • Bradford assay : 결과에 대한 data는 BSA의 농도별로 구한 absorbance를 이용 하여 standard curve를 그린 후, 식을 구해서 sample에 대한 농도를 구할 것! • Standard curve는 정확하고, 편의를 의해 excel을 사용하여 그릴 것. (standard graph 식과 R 2 값 표시할 것)

Further Study • Ideas for extension of this experiment • Protein Quantification : Lowry (copper and Folin-Ciocalteau. Phenol) : Bicinchoninicacid method : Bradford method (Coomassie brilliant blue G 250) : Biuretmethod (copper and tartrate) → 원리와 각 method의 adv. /disadv.
