102105 Gene Prediction formerly Gene Prediction 3 10262020





![DNA RNA protein Phenotype c. DNA Pevsner p 160 [1] Transcription [2] RNA processing DNA RNA protein Phenotype c. DNA Pevsner p 160 [1] Transcription [2] RNA processing](https://slidetodoc.com/presentation_image/0f41fdd9d43fe35bf2e54cc578217f91/image-9.jpg)








- Slides: 46

10/21/05 Gene Prediction (formerly Gene Prediction - 3) 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 1

Announcements Exam 2 - next Friday Posted online: Exam 2 Study Guide 544 Reading Assignment (2 papers) 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 2

Announcements 544 Semester Projects - Information needed: Please send email to me (or David) ddobbs@iastate. edu Briefly describe: • Your background & current grad research • Is there a problem related to your research you would like to learn more about & develop as project for this course? or • What would your ‘dream’ project be? 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 3

Announcements 2 Bioinformatics Seminars today (Fri Oct 21) 12: 10 PM BCB Faculty Seminar in E 164 Lagomarcino “Protein Networks” Bob Jernigan, BBMB & Director, Baker Center for Bioinformatics & Biological Statistics http: //www. bcb. iastate. edu/courses/BCB 691 -F 2005. html#Oct%2021 4: 10 PM GDCB Special Seminar in 1414 MBB “Integrating the Unknown-eome with Abiotic Stress Response Networks in Arabidopsis” Ron Mittler, Dept. of Biochem & Mol Biology University of Nevada, Reno 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 4

Gene Prediction & Regulation Mon - Gene structure review: Eukaryotes vs prokaryotes Wed - Regulatory regions: Promoters & enhancers Fri - Predicting genes - Predicting regulatory regions (? ) • Next week: Predicting RNA structure (mi. RNAs, too) 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 5

Reading Assignment Mount Bioinformatics • Chp 9 Gene Prediction & Regulation • pp 361 -385 Predicting Promoters • Ck Errata: http: //www. bioinformaticsonline. org/help/errata 2. html * Brown Genomes 2 (NCBI textbooks online) • Sect 9 Overview: Assembly of Transcription Initiation Complex • http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=genomes. chapter. 7002 • Sect 9. 1 -9. 3 DNA binding proteins, Transcription initiation • http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=genomes. section. 7016 * NOTE: Don’t worry about the details!! 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 6

Optional Reading Reviews: 1) Zhang MQ (2002) Computational prediction of eukaryotic proteincoding genes. Nat Rev Genet 3: 698 -709 http: //proxy. lib. iastate. edu: 2103/nrg/journal/v 3/n 9/full/nrg 890_fs. html 2) 1) Wasserman WW & Sandelin (2004) Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet 5: 276 -287 ml http: //proxy. lib. iastate. edu: 2103/nrg/journal/v 5/n 4/full/nrg 1315_fs. ht 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 7

Review last lecture: Gene Regulation (formerly Gene Prediction-2) c. DNAs & ESTs Uni. Gene Regulatory regions Eukaryotes vs prokaryotes 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 8
![DNA RNA protein Phenotype c DNA Pevsner p 160 1 Transcription 2 RNA processing DNA RNA protein Phenotype c. DNA Pevsner p 160 [1] Transcription [2] RNA processing](https://slidetodoc.com/presentation_image/0f41fdd9d43fe35bf2e54cc578217f91/image-9.jpg)
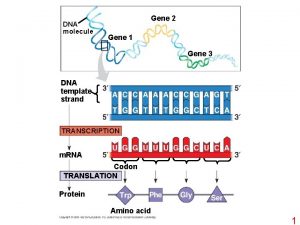
DNA RNA protein Phenotype c. DNA Pevsner p 160 [1] Transcription [2] RNA processing (splicing) [3] RNA export [4] RNA surveillance 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 9

Uni. Gene: unique genes via ESTs • Find Uni. Gene at NCBI: www. ncbi. nlm. nih. gov/Uni. Gene • Uni. Gene clusters contain many ESTs • Uni. Gene data come from many c. DNA libraries. Thus, when you look up a gene in Uni. Gene you get information on its abundance and its regional distribution Pevsner p 164 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 10

Today: Gene Prediction (formerly Gene Prediction - 3) Predicting genes Mon - Predicting regulatory regions Focus on promoters Introduction to RNA Later: Genome browsers 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 11

Gene Prediction • Overview of steps & strategies • What sequence signals can be used? • What other types of information can be used? • Algorithms • HMMs, discriminant functions, neural nets • Gene prediction software • 3 major types • many, many programs! 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 12

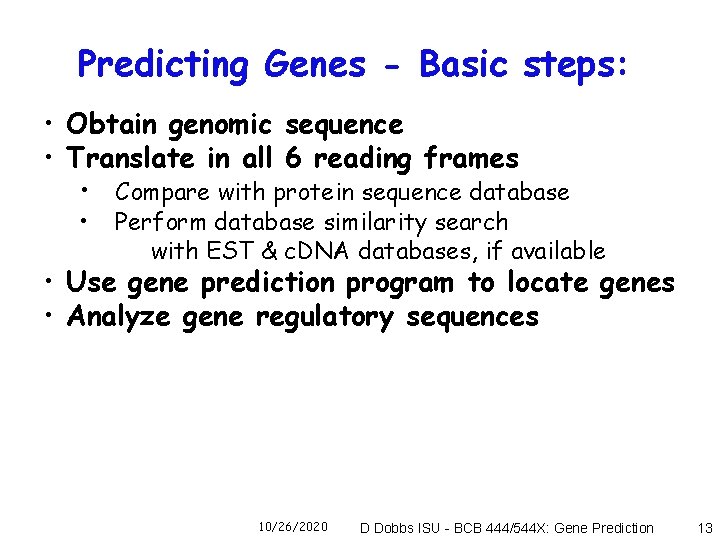
Predicting Genes - Basic steps: • Obtain genomic sequence • Translate in all 6 reading frames • Compare with protein sequence database • Perform database similarity search with EST & c. DNA databases, if available • Use gene prediction program to locate genes • Analyze gene regulatory sequences 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 13

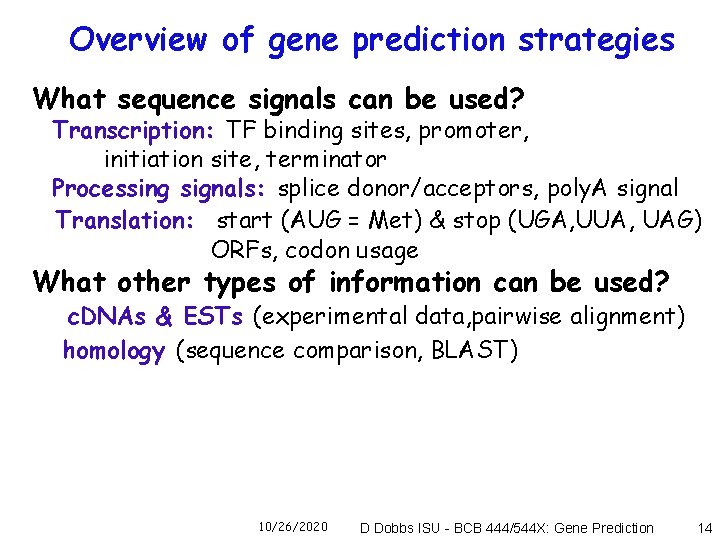
Overview of gene prediction strategies What sequence signals can be used? Transcription: TF binding sites, promoter, initiation site, terminator Processing signals: splice donor/acceptors, poly. A signal Translation: start (AUG = Met) & stop (UGA, UUA, UAG) ORFs, codon usage What other types of information can be used? c. DNAs & ESTs (experimental data, pairwise alignment) homology (sequence comparison, BLAST) 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 14

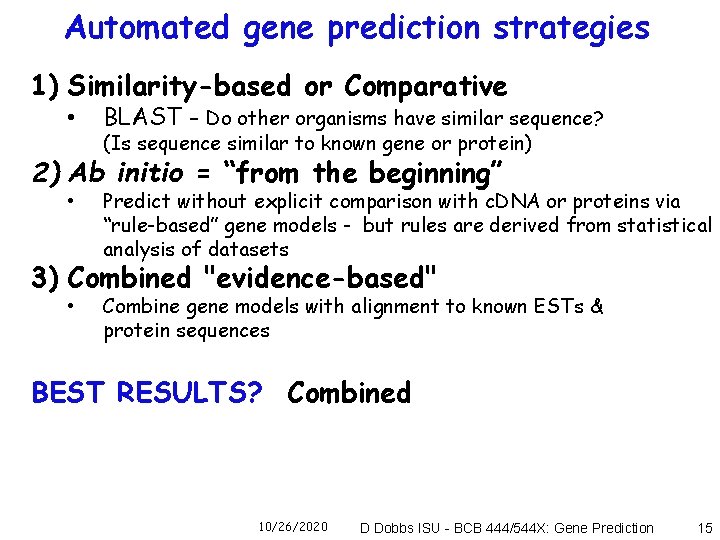
Automated gene prediction strategies 1) Similarity-based or Comparative • BLAST - Do other organisms have similar sequence? (Is sequence similar to known gene or protein) 2) Ab initio = “from the beginning” • Predict without explicit comparison with c. DNA or proteins via “rule-based” gene models - but rules are derived from statistical analysis of datasets 3) Combined "evidence-based" • Combine gene models with alignment to known ESTs & protein sequences BEST RESULTS? Combined 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 15

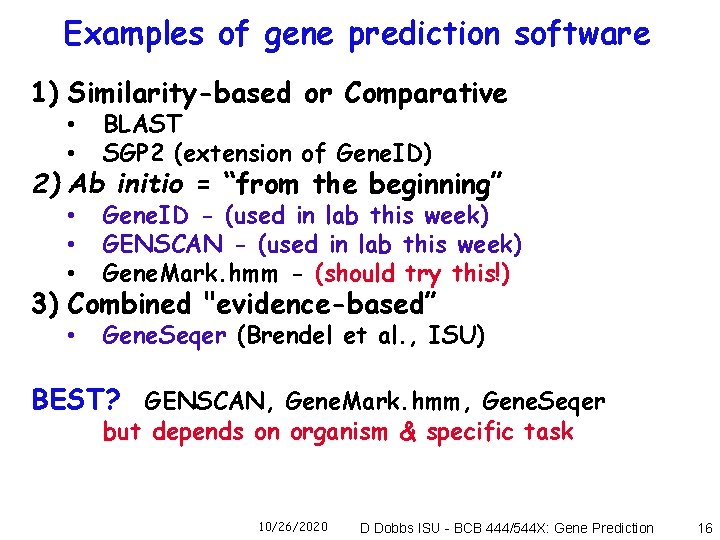
Examples of gene prediction software 1) Similarity-based or Comparative • • BLAST SGP 2 (extension of Gene. ID) • • • Gene. ID - (used in lab this week) GENSCAN - (used in lab this week) Gene. Mark. hmm - (should try this!) • Gene. Seqer (Brendel et al. , ISU) 2) Ab initio = “from the beginning” 3) Combined "evidence-based” BEST? GENSCAN, Gene. Mark. hmm, Gene. Seqer but depends on organism & specific task 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 16

Gene prediction: Eukaryotes vs prokaryotes Gene prediction is easier in microbial genomes Why? Smaller genomes Simpler gene structures More sequenced genomes! (for comparative approaches) Methods? Previously, mostly HMM-based Now: similarity-based methods because so many genomes available 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 17

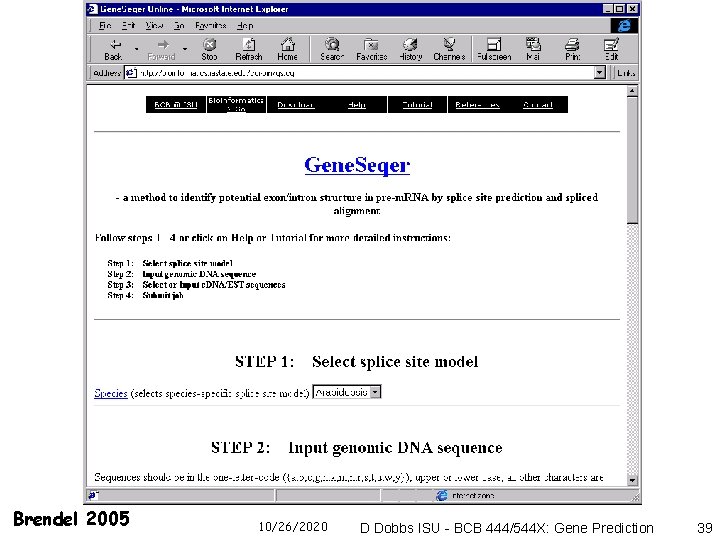
Gene. Seqer - Brendel et al. http: //deepc 2. psi. iastate. edu/cgi-bin/gs. cgi 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 18

Thanks to Volker Brendel, ISU for following Figs & Slides Slightly modified from: BSSI Genome Informatics Module http: //www. bioinformatics. iastate. edu/BBSI/course_de sc_2005. html#module. B V Brendel vbrendel@iastate. edu 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 19

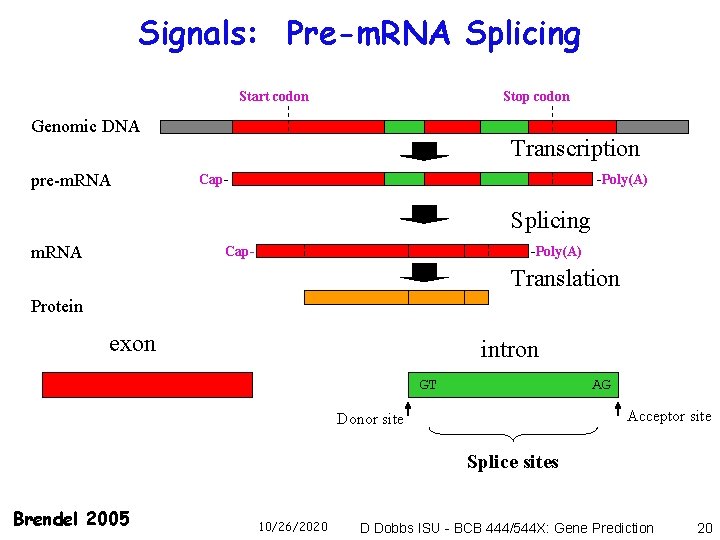
Signals: Pre-m. RNA Splicing Start codon Stop codon Genomic DNA pre-m. RNA Transcription Cap- -Poly(A) Splicing m. RNA -Poly(A) Cap- Translation Protein exon intron GT AG Acceptor site Donor site Splice sites Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 20

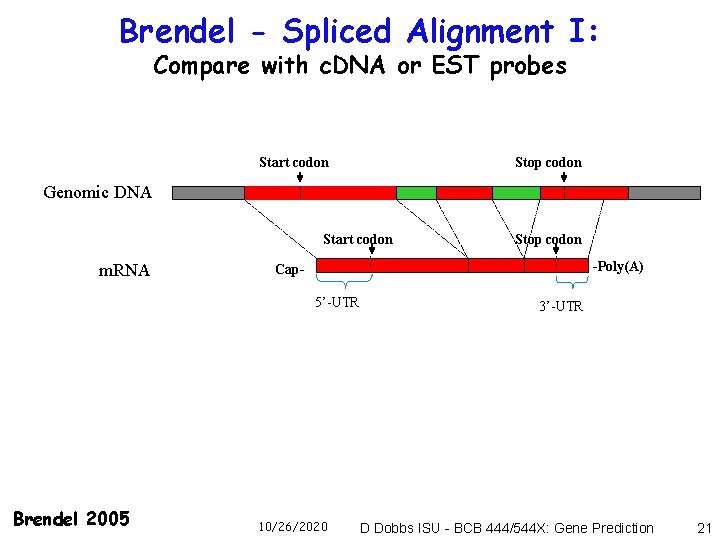
Brendel - Spliced Alignment I: Compare with c. DNA or EST probes Start codon Stop codon Genomic DNA Start codon m. RNA -Poly(A) Cap 5’-UTR Brendel 2005 Stop codon 10/26/2020 3’-UTR D Dobbs ISU - BCB 444/544 X: Gene Prediction 21

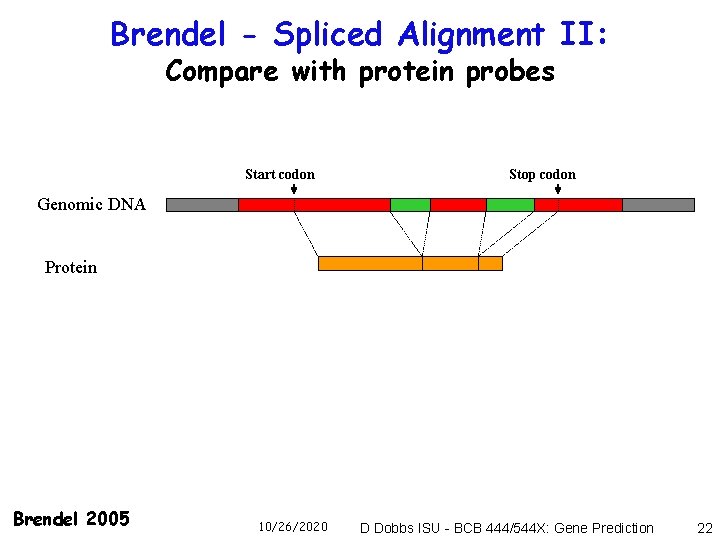
Brendel - Spliced Alignment II: Compare with protein probes Start codon Stop codon Genomic DNA Protein Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 22

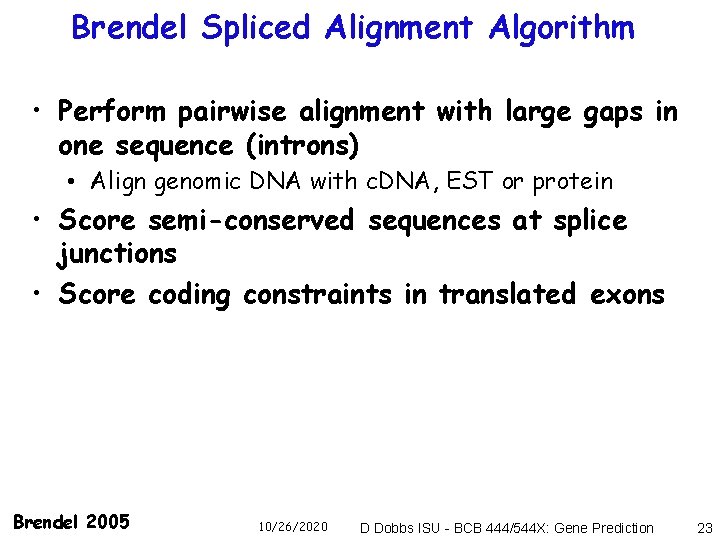
Brendel Spliced Alignment Algorithm • Perform pairwise alignment with large gaps in one sequence (introns) • Align genomic DNA with c. DNA, EST or protein • Score semi-conserved sequences at splice junctions • Score coding constraints in translated exons Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 23

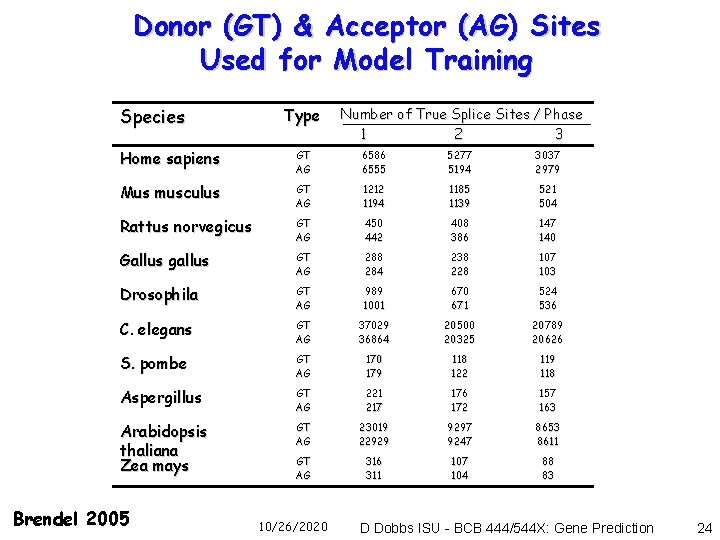
Donor (GT) & Acceptor (AG) Sites Used for Model Training Species Type Number of True Splice Sites / Phase 1 2 3 Home sapiens GT AG 6586 6555 5277 5194 3037 2979 Mus musculus GT AG 1212 1194 1185 1139 521 504 Rattus norvegicus GT AG 450 442 408 386 147 140 Gallus gallus GT AG 288 284 238 228 107 103 Drosophila GT AG 989 1001 670 671 524 536 C. elegans GT AG 37029 36864 20500 20325 20789 20626 S. pombe GT AG 170 179 118 122 119 118 Aspergillus GT AG 221 217 176 172 157 163 Arabidopsis thaliana Zea mays GT AG 23019 22929 9297 9247 8653 8611 GT AG 316 311 107 104 88 83 Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 24

Splice Site Detection • Information Content Ii : • Extent of Splice Signal Window: i : ith position in sequence Ī : average information content over all positions i > 20 nt from splice site Ī : average standard deviation of Ī Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 25

Results? Brendel 2005 Human T 2_GT Human T 2_AG Human F 1_AG Human Fi_AG A. thaliana T 2_GT A. thaliana T 2_AG A. thaliana F 1_AG A. thaliana Fi_AG 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 26

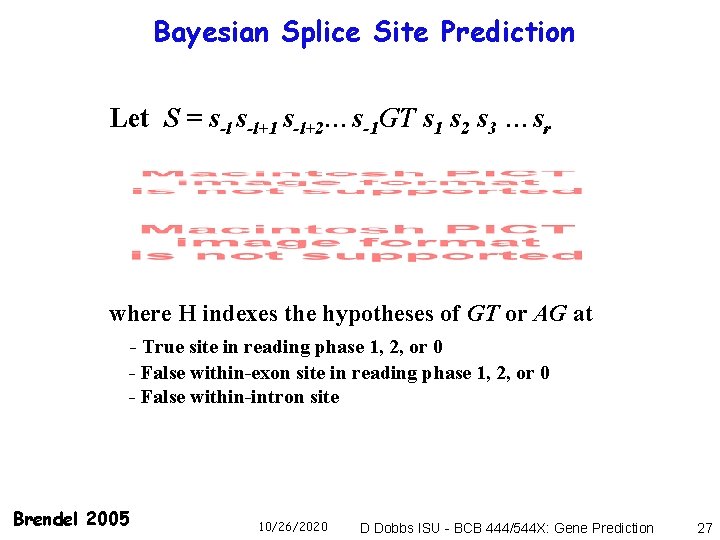
Bayesian Splice Site Prediction Let S = s-l+1 s-l+2…s-1 GT s 1 s 2 s 3 …sr where H indexes the hypotheses of GT or AG at - True site in reading phase 1, 2, or 0 - False within-exon site in reading phase 1, 2, or 0 - False within-intron site Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 27

Bayes Factor as Decision Criterion H 0: H=T: - 2 -class model: - 7 class model: Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 28

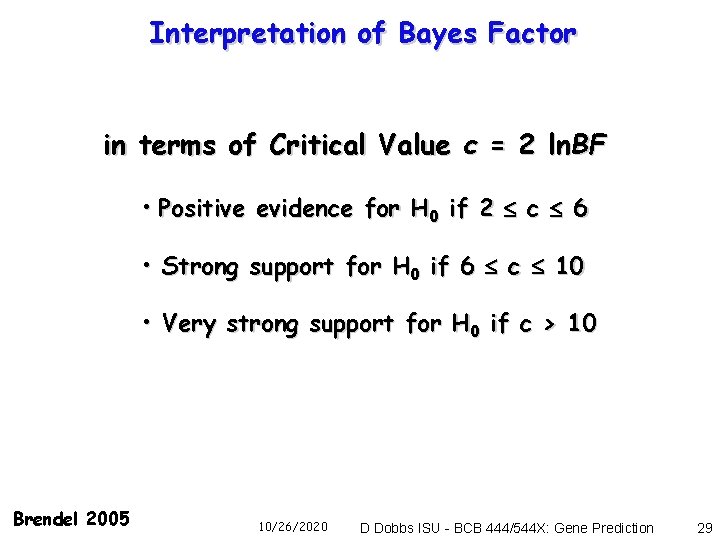
Interpretation of Bayes Factor in terms of Critical Value c = 2 ln. BF • Positive evidence for H 0 if 2 c 6 • Strong support for H 0 if 6 c 10 • Very strong support for H 0 if c > 10 Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 29

Evaluation of Splice Site Prediction Actual True False Predicted True TP FP PP=TP+FP False FN TN PN=FN+TN AP=TP+FN AN=FP+TN = Coverage • Sensitivity: • Specificity: • Misclassification rates: • Normalized specificity: Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 30

Species Homo sapiens Drosophila C. elegans A. thaliana Brendel 2005 Model 2 C 2 C 7 C 7 C Site Test Site Set True False GT 921 44411 AG 920 65103 GT 329 11501 AG 329 14920 GT 400 7460 AG 400 10132 GT 613 9027 AG 614 10196 10/26/2020 Bayes Factor Sn Sp (%) (%) 0 3 6 98. 5 91. 7 66. 3 90. 3 76. 1 90. 5 96. 3 98. 5 88. 4 92. 9 96. 1 16. 4 34. 8 57. 6 9. 7 15. 7 25. 6 0 3 6 95. 4 90. 0 83. 9 95. 7 92. 1 85. 1 94. 8 97. 6 99. 1 94. 8 97. 0 98. 5 34. 1 53. 6 75. 0 28. 7 41. 4 59. 4 0 3 6 97. 8 94. 2 84. 8 98. 8 96. 2 90. 2 92. 7 97. 1 99. 1 97. 2 98. 8 99. 5 40. 4 64. 3 85. 4 58. 2 76. 9 88. 5 0 3 6 99. 5 95. 6 87. 1 99. 2 96. 4 87. 1 93. 2 97. 6 99. 3 92. 3 96. 4 98. 6 48. 1 73. 2 91. 0 41. 9 62. 0 81. 2 D Dobbs ISU - BCB 444/544 X: Gene Prediction 31

Performance? Human GT site Human AG site Sn Sn C. elegans GT site C. elegans AG site Sn Sn A. thaliana GT site Sn Brendel 2005 10/26/2020 A. thaliana AG site Sn D Dobbs ISU - BCB 444/544 X: Gene Prediction 32

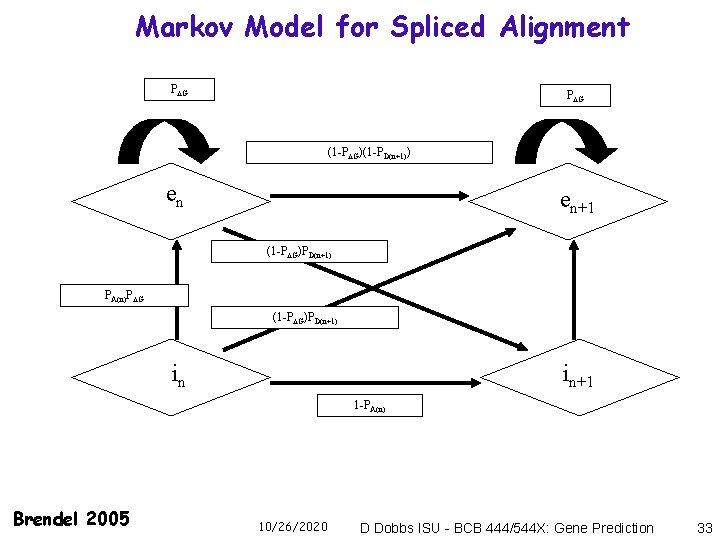
Markov Model for Spliced Alignment P G (1 -P G)(1 -PD(n+1)) en en+1 (1 -P G)PD(n+1) PA(n)P G (1 -P G)PD(n+1) in in+1 1 -PA(n) Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 33

Performance vs other methods • Comparison with ab initio gene prediction programs? • Depends on: • Availability of ESTs • Availability of protein homologs Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 34

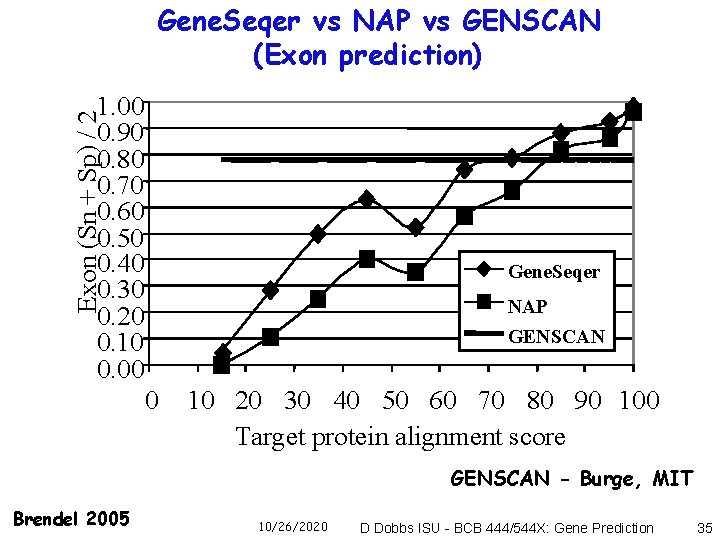
Gene. Seqer vs NAP vs GENSCAN (Exon prediction) Exon (Sn + Sp) / 2 1. 00 0. 90 0. 80 0. 70 0. 60 0. 50 0. 40 0. 30 0. 20 0. 10 0. 00 Gene. Seqer NAP GENSCAN 0 10 20 30 40 50 60 70 80 90 100 Target protein alignment score GENSCAN - Burge, MIT Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 35

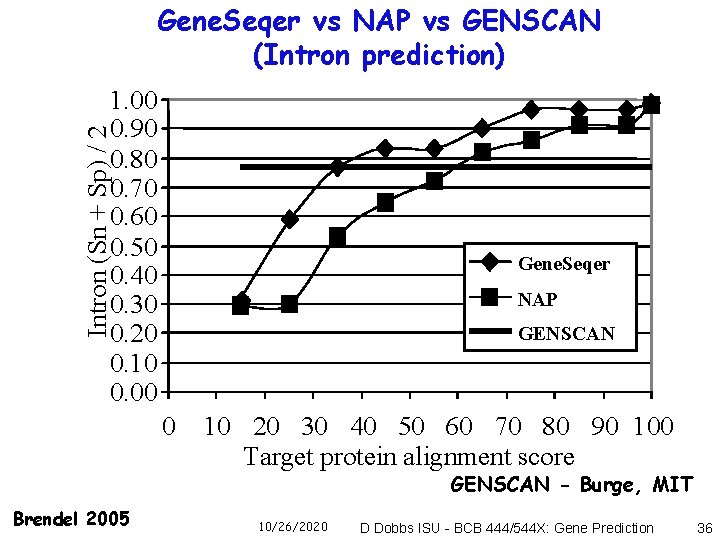
Gene. Seqer vs NAP vs GENSCAN (Intron prediction) Intron (Sn + Sp) / 2 1. 00 0. 90 0. 80 0. 70 0. 60 0. 50 0. 40 0. 30 0. 20 0. 10 0. 00 Gene. Seqer NAP GENSCAN 0 10 20 30 40 50 60 70 80 90 100 Target protein alignment score GENSCAN - Burge, MIT Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 36

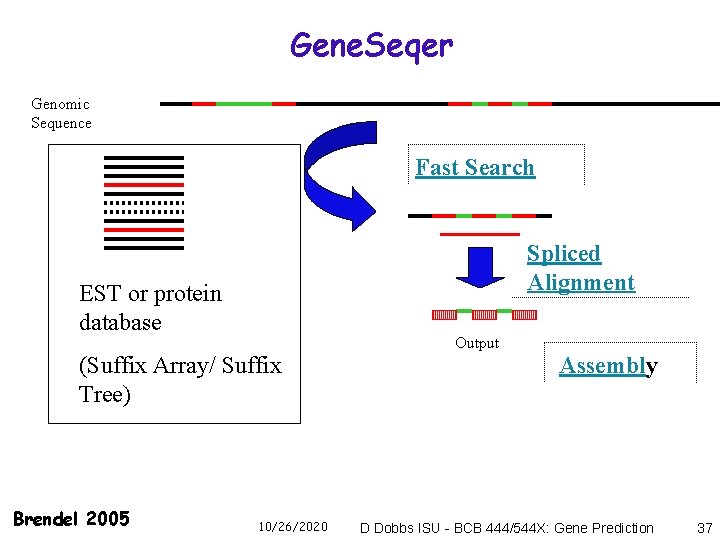
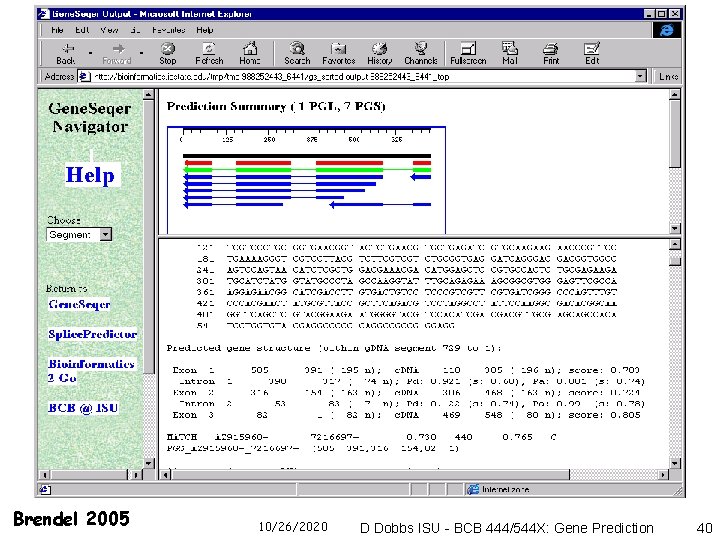
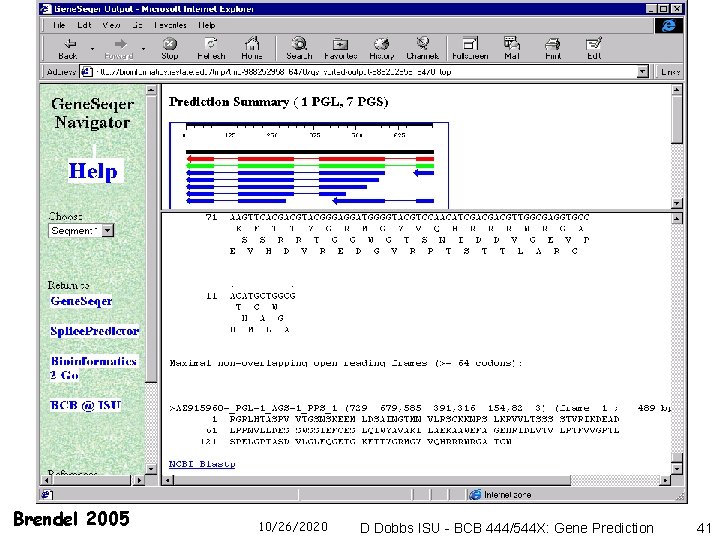
Gene. Seqer Genomic Sequence Fast Search Spliced Alignment EST or protein database Output (Suffix Array/ Suffix Tree) Brendel 2005 10/26/2020 Assembly D Dobbs ISU - BCB 444/544 X: Gene Prediction 37

Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 38

Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 39

Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 40

Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 41

Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 42

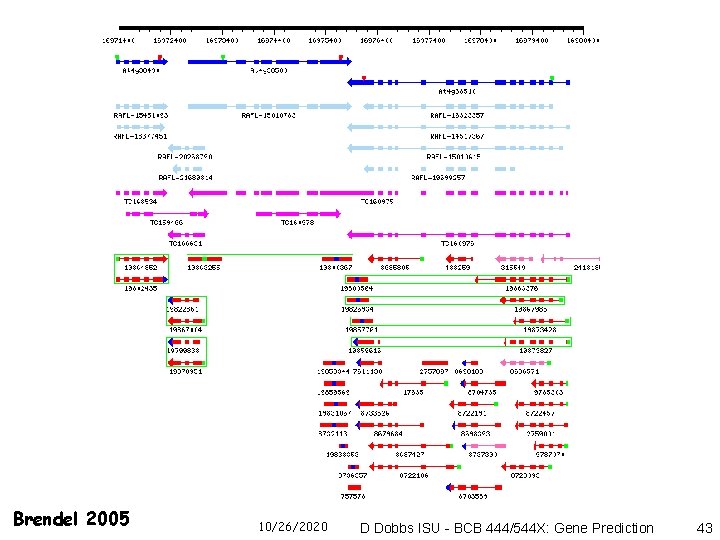
Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 43

Gene Structure Annotation - Problems False positive intergenic region: • 2 annotated genes actually correspond to a single gene False negative intergenic region: • One annotated gene structure actually contains 2 genes False negative gene prediction: • Missing gene (no annotation) Other: • partially incorrect gene annotation • missing annotation of alternative transcripts Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 44

Brendel 2005 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 45

Other Resources Current Protocols in Bioinformatics http: //www. 4 ulr. com/products/currentprotocols/bioinformatics. html Finding Genes 4. 1 An Overview of Gene Identification: Approaches, Strategies, and Considerations 4. 2 Using MZEF To Find Internal Coding Exons 4. 3 Using GENEID to Identify Genes 4. 4 Using Glimmer. M to Find Genes in Eukaryotic Genomes 4. 5 Prokaryotic Gene Prediction Using Gene. Mark and Gene. Mark. hmm 4. 6 Eukaryotic Gene Prediction Using Gene. Mark. hmm 4. 7 Application of First. EF to Find Promoters and First Exons in the Human Genome 4. 8 Using TWINSCAN to Predict Gene Structures in Genomic DNA Sequences 4. 9 Grail. EXP and Genome Analysis Pipeline for Genome Annotation 4. 10 Using Repeat. Masker to Identify Repetitive Elements in Genomic Sequences 10/26/2020 D Dobbs ISU - BCB 444/544 X: Gene Prediction 46
 Formerly restricted data
Formerly restricted data Absence management formerly aesop
Absence management formerly aesop Bank alfalah formerly
Bank alfalah formerly Sxole
Sxole Gene prediction in prokaryotes and eukaryotes
Gene prediction in prokaryotes and eukaryotes Chapter 17: from gene to protein
Chapter 17: from gene to protein Gene by gene test results
Gene by gene test results Correlation vs prediction
Correlation vs prediction A testable prediction
A testable prediction Solar cycle 25 prediction
Solar cycle 25 prediction Tournament branch predictor
Tournament branch predictor Social learning theory julian rotter
Social learning theory julian rotter Revenue prediction machine learning
Revenue prediction machine learning Genetic algorithm for stock prediction
Genetic algorithm for stock prediction Masta prediction
Masta prediction Prediction format
Prediction format Prediction error variance
Prediction error variance Branch prediction
Branch prediction Good readers making predictions by
Good readers making predictions by Derition
Derition Hunger games chapter 8 questions and answers
Hunger games chapter 8 questions and answers Meritsprediction
Meritsprediction Perceptual linear prediction
Perceptual linear prediction Hypothesis vs prediction
Hypothesis vs prediction Prediction vs extrapolation
Prediction vs extrapolation Prediction pip
Prediction pip Ncash prediction
Ncash prediction How to calculate sst in regression
How to calculate sst in regression Andini prediction
Andini prediction Regional atmospheric soaring prediction
Regional atmospheric soaring prediction Punnet square eye color
Punnet square eye color Iterative prediction of motion
Iterative prediction of motion Prediction vs inference venn diagram
Prediction vs inference venn diagram Branch prediction
Branch prediction Hypothesis vs prediction
Hypothesis vs prediction Freedico prediction
Freedico prediction Prediction of nba games based on machine learning methods
Prediction of nba games based on machine learning methods Hunger games questions by chapter
Hunger games questions by chapter Micosis
Micosis Making inferences and predictions
Making inferences and predictions Mathematical models for impact prediction
Mathematical models for impact prediction Branch prediction techniques
Branch prediction techniques Tipsy prediction
Tipsy prediction Tournament prediction computer architecture
Tournament prediction computer architecture Drag prediction workshop
Drag prediction workshop Branch prediction in computer architecture
Branch prediction in computer architecture Branch predictor btb
Branch predictor btb