10192021 Master title Molecular Interactions the Int Act




- Slides: 72

10/19/2021 Master title Molecular Interactions – the Int. Act Database 5 Sandra Orchard EMBL-EBI is an Outstation of the European Molecular Biology Laboratory.

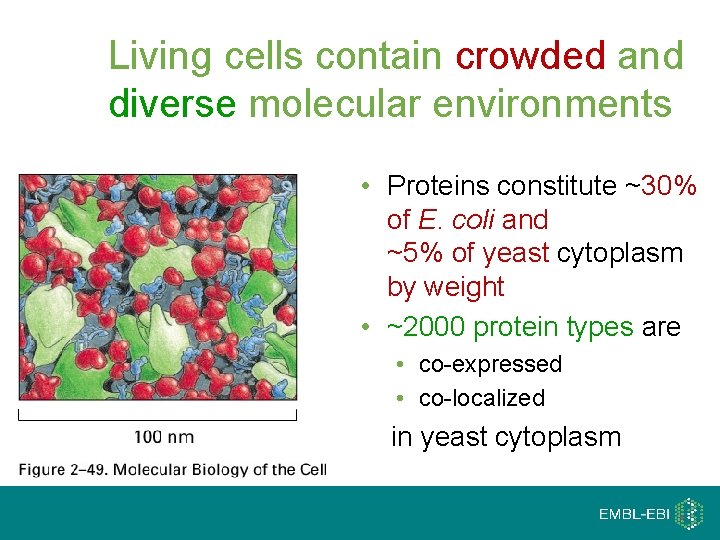
Living cells contain crowded and diverse molecular environments • Proteins constitute ~30% of E. coli and ~5% of yeast cytoplasm by weight • ~2000 protein types are • co-expressed • co-localized in yeast cytoplasm

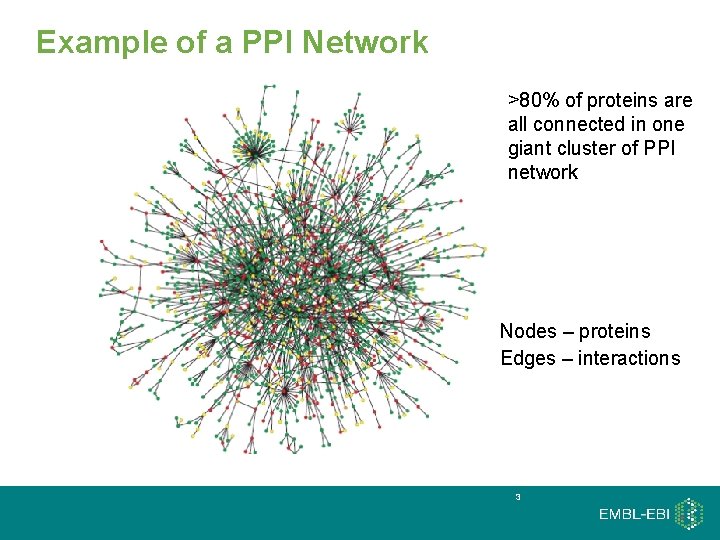
Example of a PPI Network >80% of proteins are all connected in one giant cluster of PPI network • Nodes – proteins • Edges – interactions 3

Why is it useful to study PPI networks? • Proteins are the workhorses of cell, carrying out catalytic reactions, transport, forming viral caspids, transmitting information from DNA to RNA, traverse membranes, forming regulated channel, make possible synthesis of new proteins, responsible for degradation of unnecessary proteins, vehicles of immune response • One way to predict protein function is through identification of binding partners – Guilt by Association • If the function of at least one of the components with which the protein interacts is known, that should let us assign its function(s) and the pathway(s) • Hence, through the intricate network of these interactions we can map cellular pathways, their interconnectivities and their dynamic regulation 4

Why is it useful to study the structure of PPI networks? • Common properties of biological networks • Can help us relate network structure to biological function • Protein’s relative position in a network • Correlate conserved functional modules with protein complexes 5

Int. Act goals & achievements 1. Publicly available repository of molecular interactions (mainly PPIs) - ~300 K binary interactions taken from >5, 300 publications (May 2012) 2. Data is standards-compliant and available via our website, for download at our ftp site or via PSICQUIC http: //www. ebi. ac. uk/intact ftp: //ftp. ebi. ac. uk/pub/databases/intact www. ebi. ac. uk/Tools/webservices/psicquic/view/main. xhtml 3. Provide open-access versions of the software to allow installation of local Int. Act nodes. 6

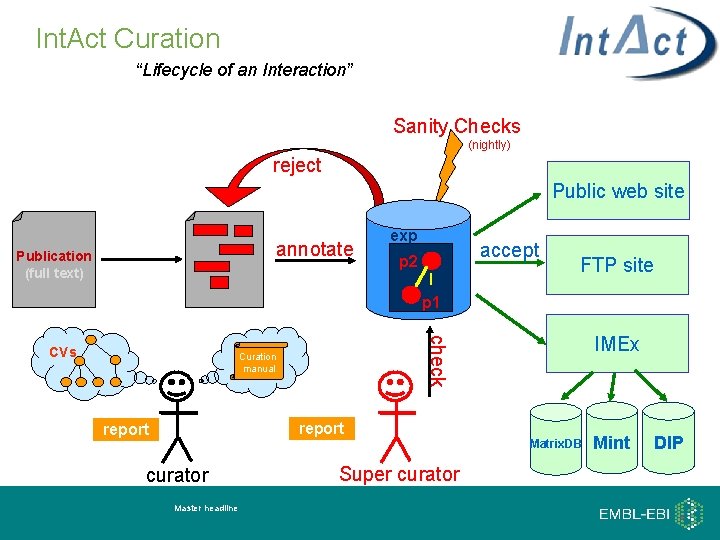
Int. Act Curation “Lifecycle of an Interaction” Sanity Checks (nightly) reject Public web site Publication (full text) . exp accept p 2 I p 1 Curation manual report curator Master headline FTP site IMEx check CVs annotate Super curator Matrix. DB Mint DIP

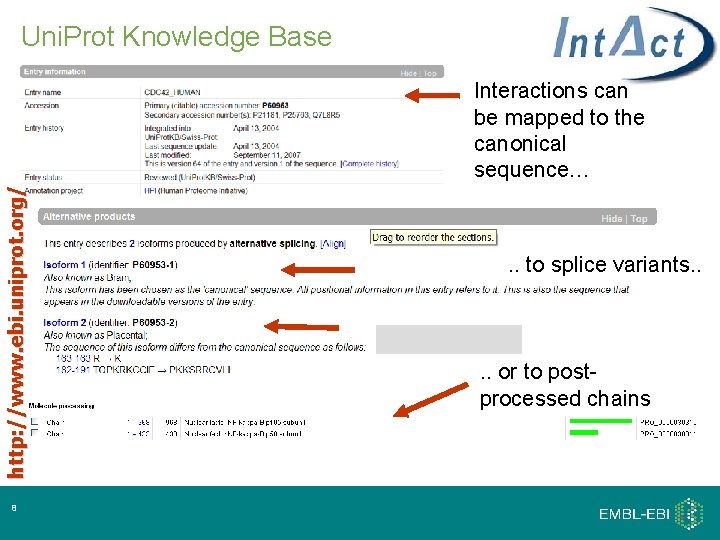
Uni. Prot Knowledge Base http: //www. ebi. uniprot. org/ Interactions can be mapped to the canonical sequence… 8 . . to splice variants. . or to postprocessed chains

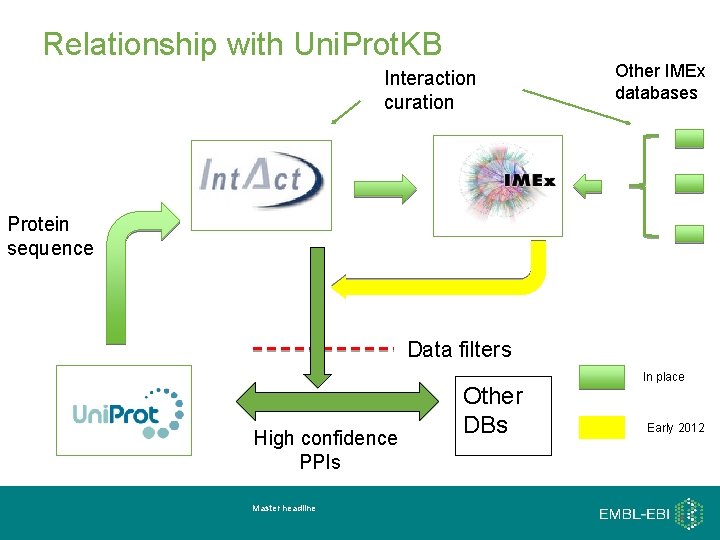
Relationship with Uni. Prot. KB Interaction curation Other IMEx databases Protein sequence Data filters High confidence PPIs Master headline Other DBs In place Early 2012

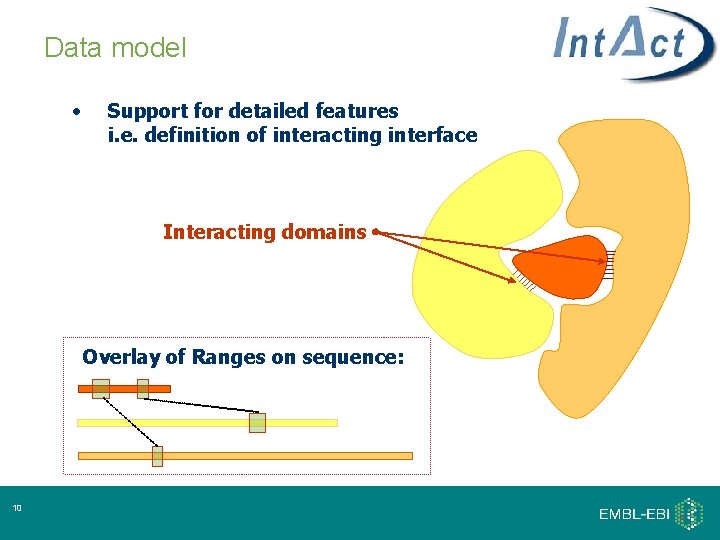
Data model • Support for detailed features i. e. definition of interacting interface Interacting domains Overlay of Ranges on sequence: 10

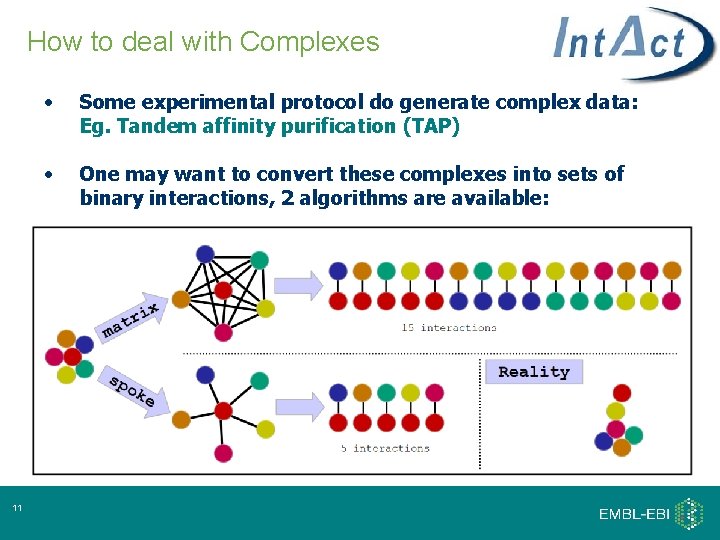
How to deal with Complexes 11 • Some experimental protocol do generate complex data: Eg. Tandem affinity purification (TAP) • One may want to convert these complexes into sets of binary interactions, 2 algorithms are available:

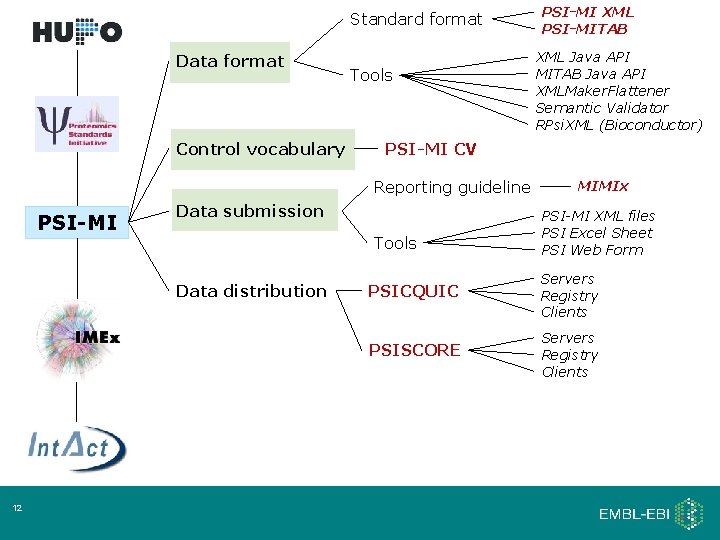
Standard format Data format Control vocabulary Tools Data submission Tools Data distribution 12 XML Java API MITAB Java API XMLMaker. Flattener Semantic Validator RPsi. XML (Bioconductor) PSI-MI CV Reporting guideline PSI-MI XML PSI-MITAB MIMIx PSI-MI XML files PSI Excel Sheet PSI Web Form PSICQUIC Servers Registry Clients PSISCORE Servers Registry Clients

Performing and visualing a Simple Search Data, Standards and Tools EBI Walthrough May 2009 EBI is an Outstation of the European Molecular Biology Laboratory.

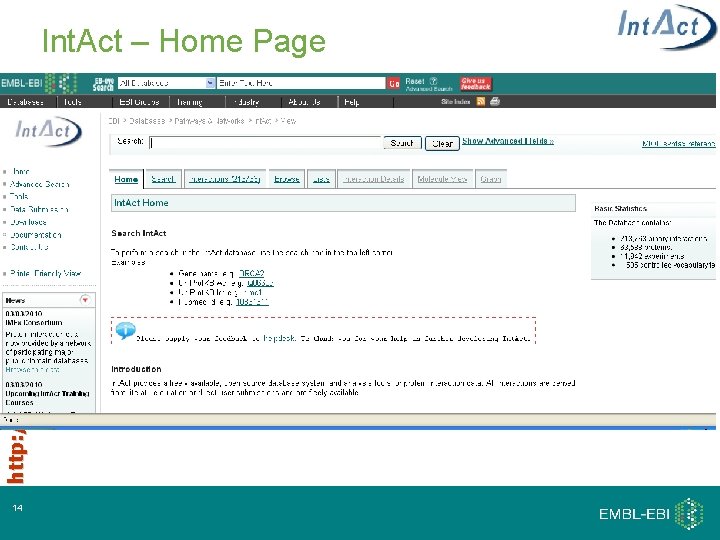
http: //www. ebi. ac. uk/intact Int. Act – Home Page 14

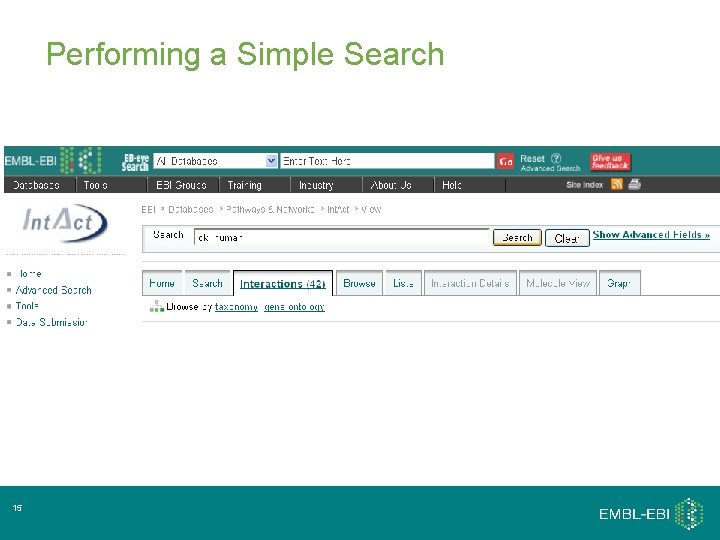
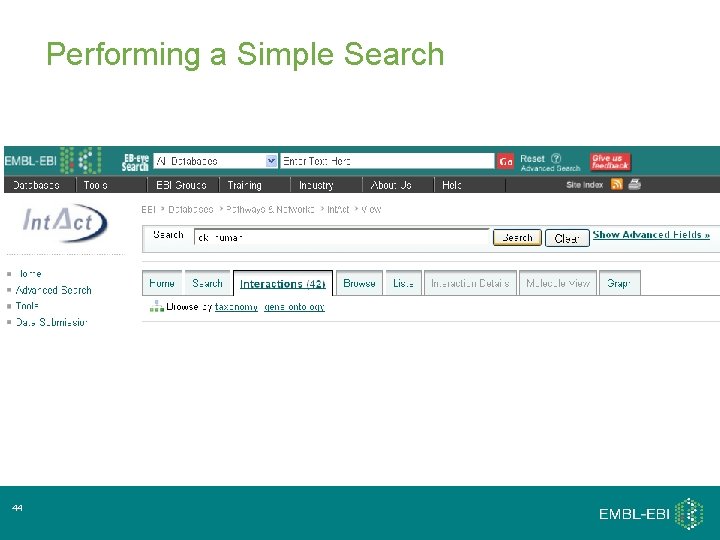
Performing a Simple Search 15

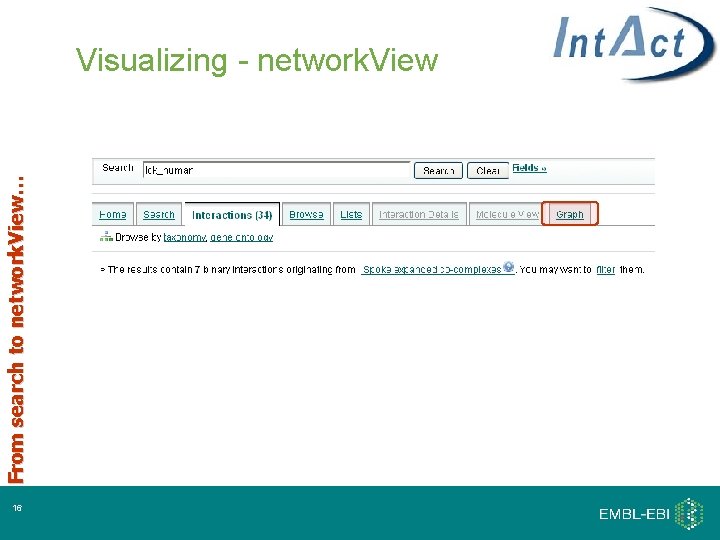
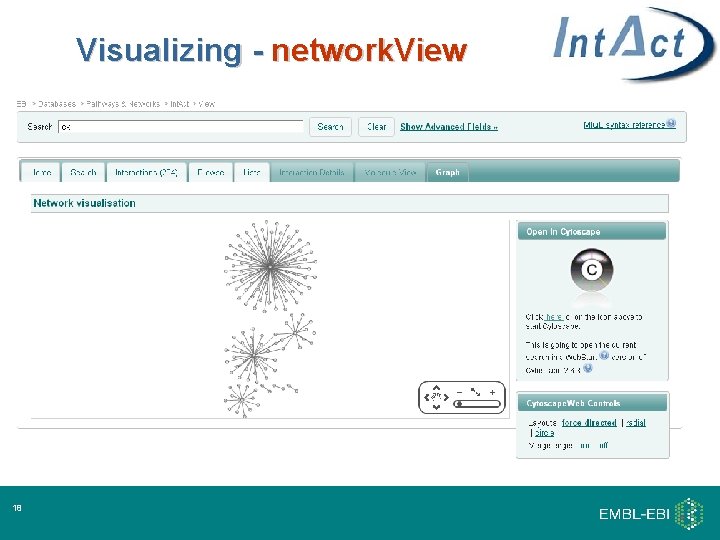
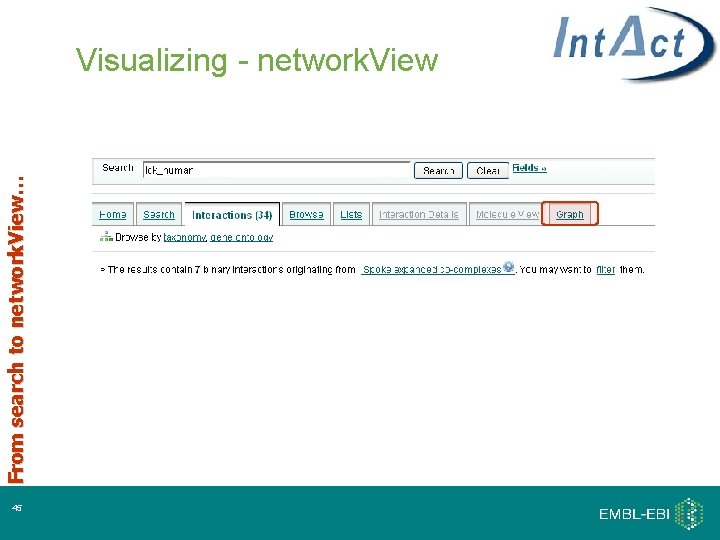
From search to network. View… Visualizing - network. View 16

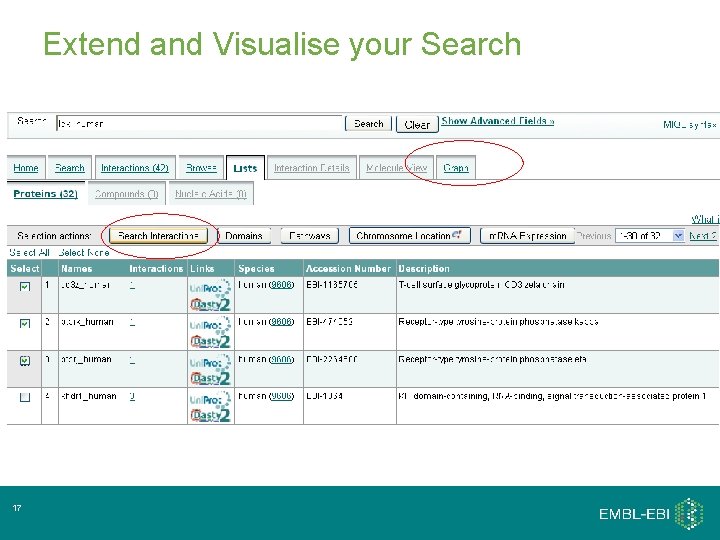
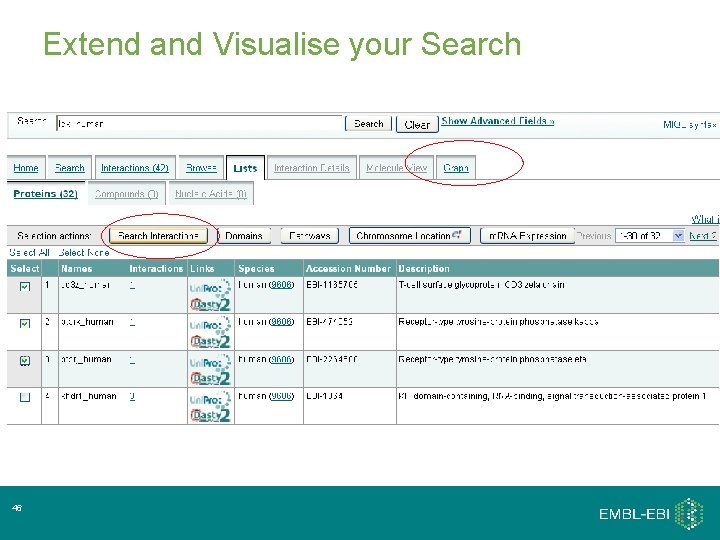
Extend and Visualise your Search 17

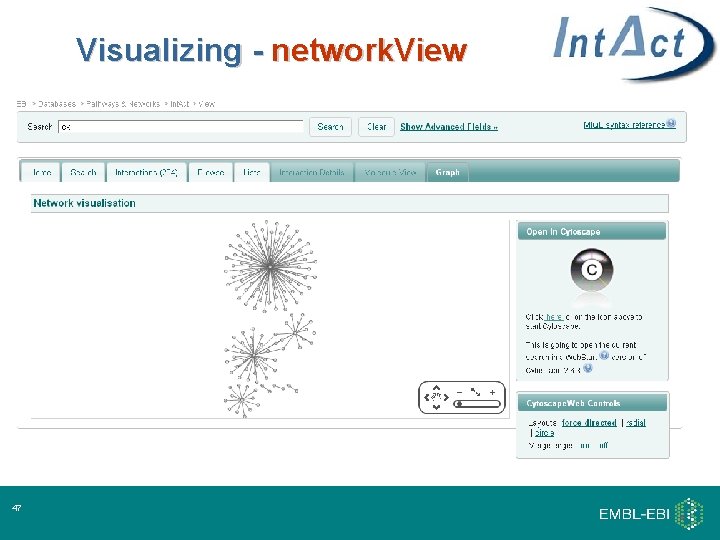
Visualizing - network. View 18

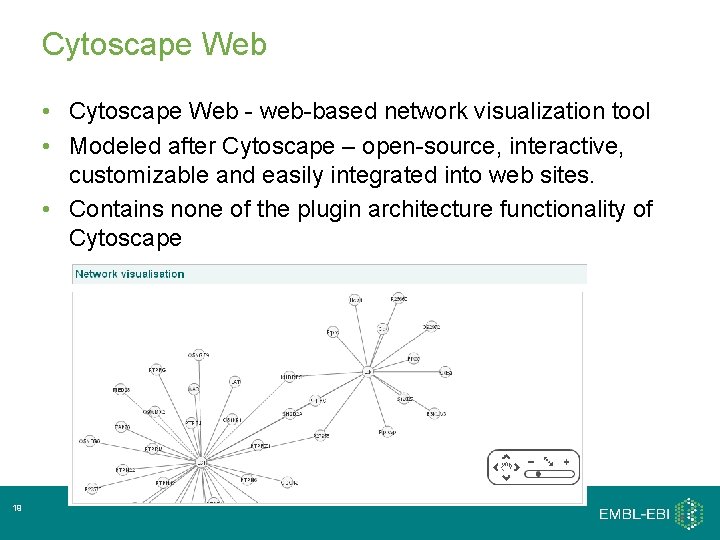
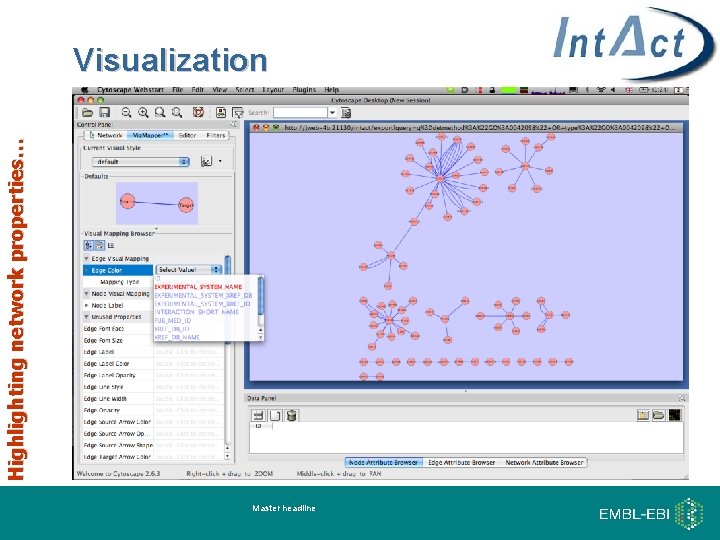
Cytoscape Web • Cytoscape Web - web-based network visualization tool • Modeled after Cytoscape – open-source, interactive, customizable and easily integrated into web sites. • Contains none of the plugin architecture functionality of Cytoscape 19

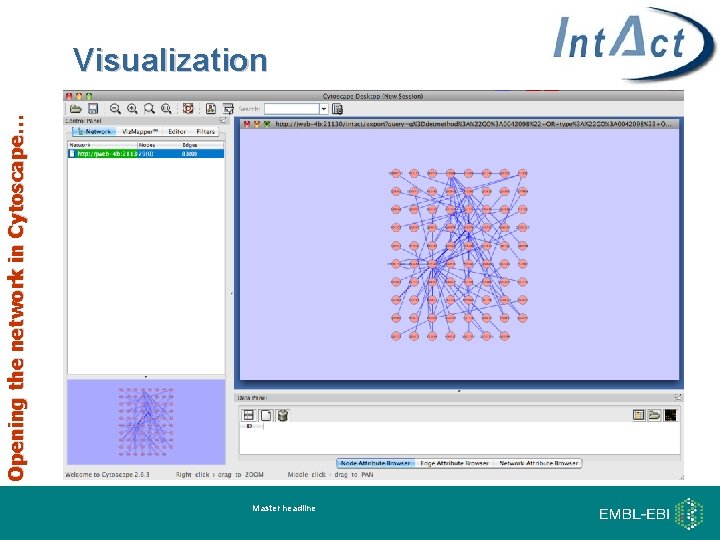
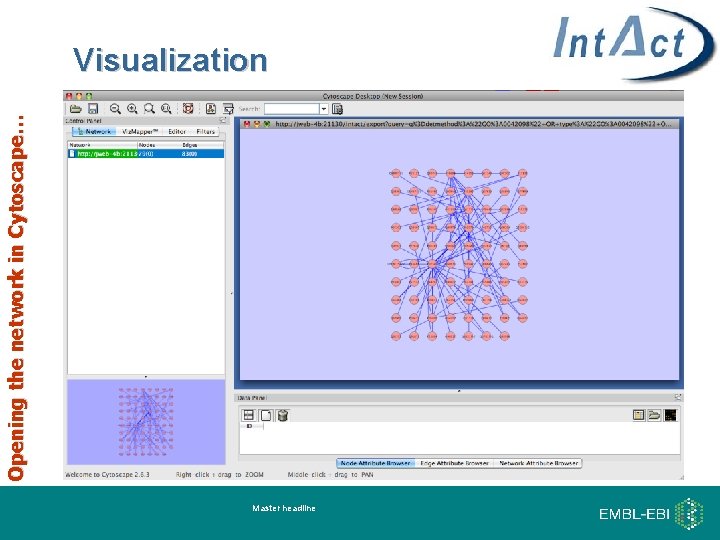
Opening the network in Cytoscape… Visualization Master headline

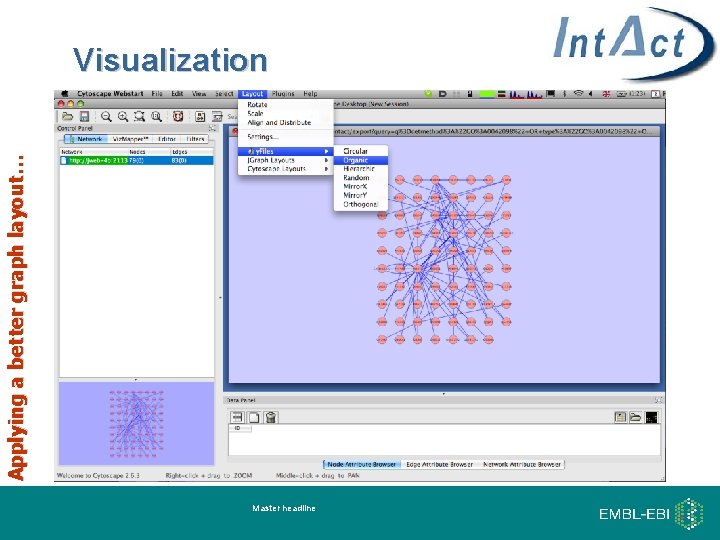
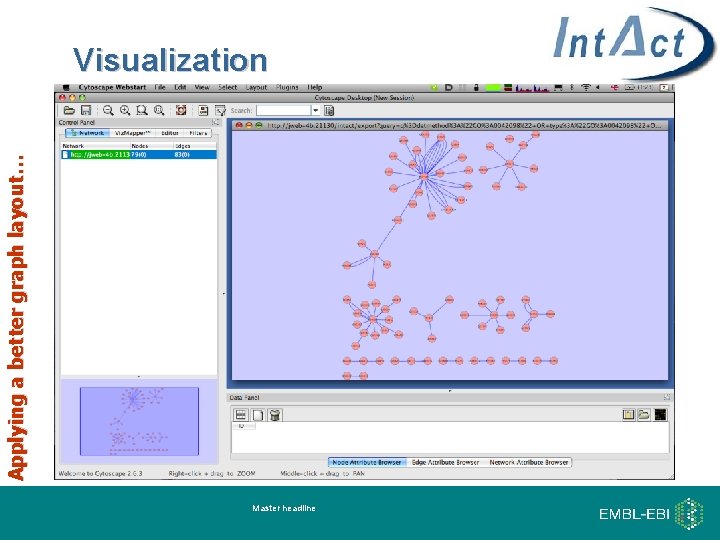
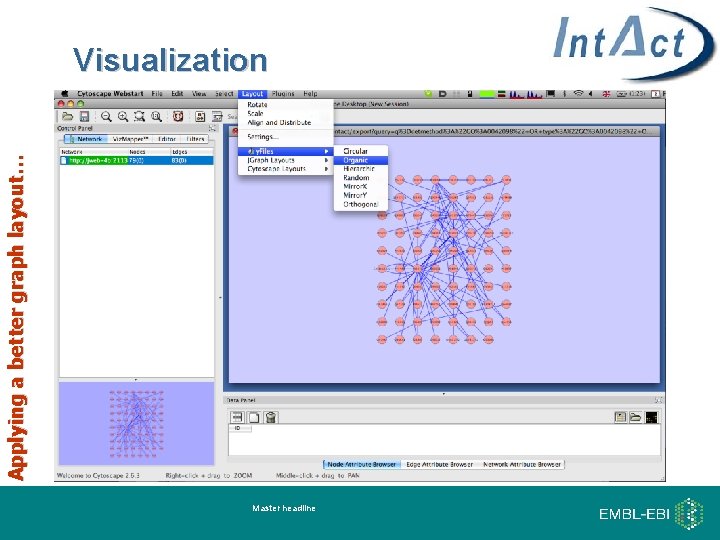
Applying a better graph layout… Visualization Master headline

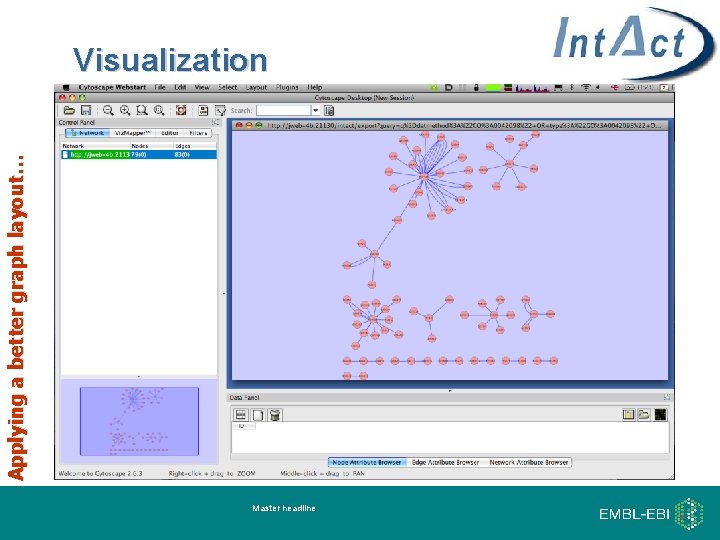
Applying a better graph layout… Visualization Master headline

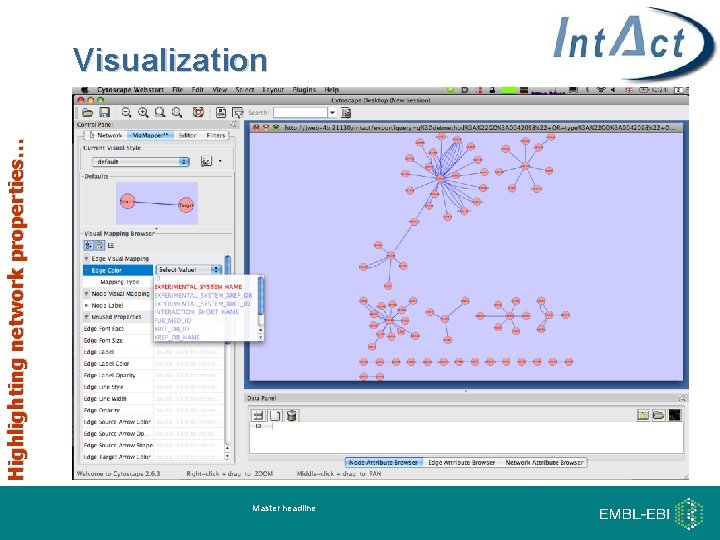
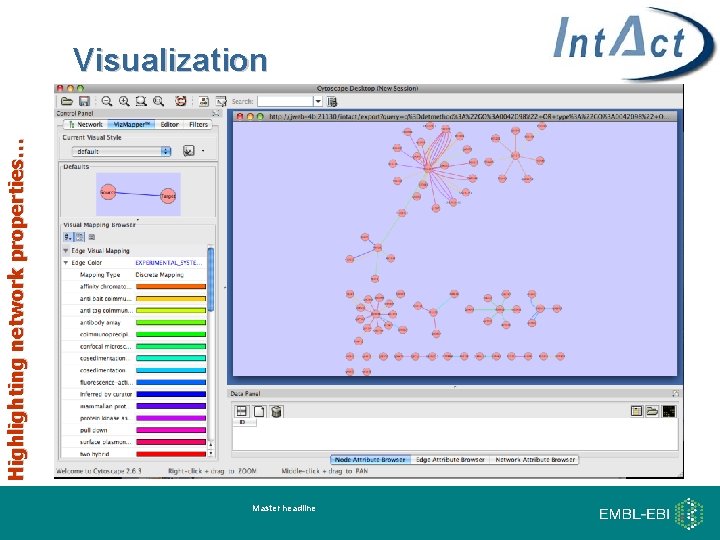
Highlighting network properties… Visualization Master headline

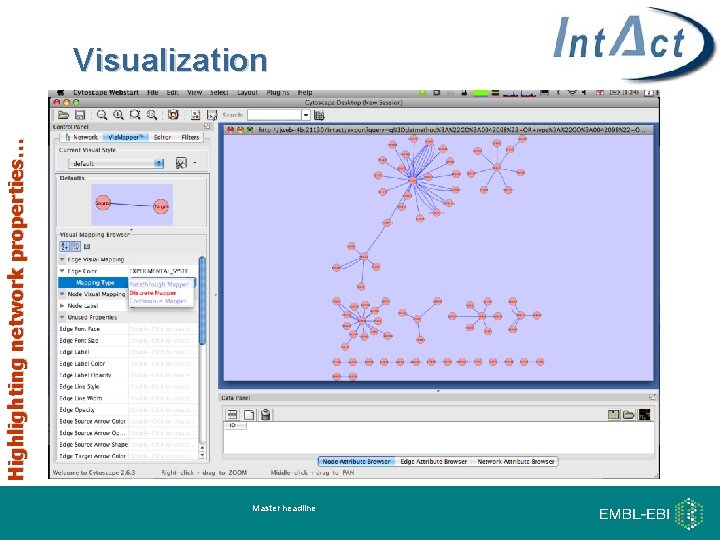
Highlighting network properties… Visualization Master headline

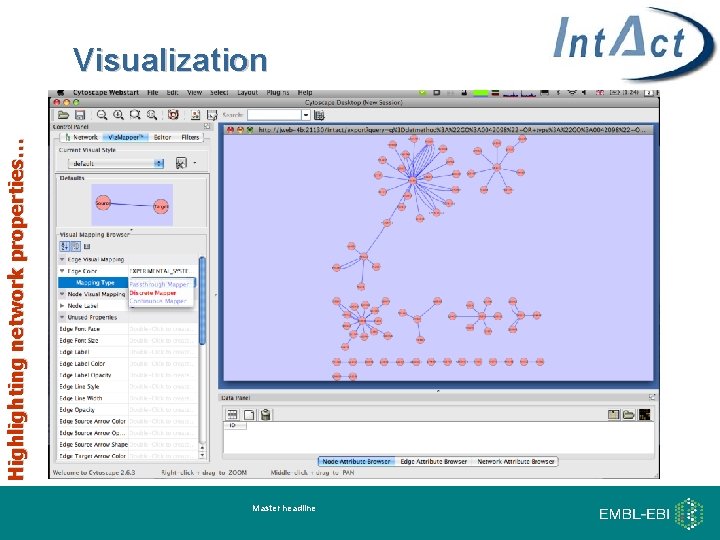
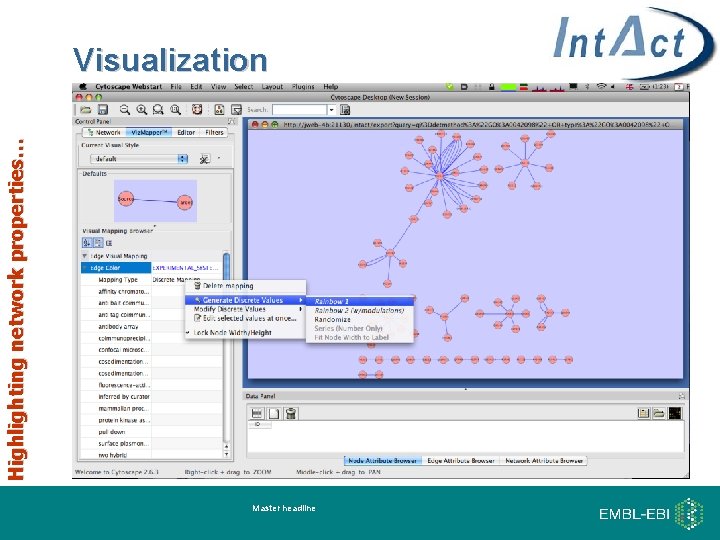
Highlighting network properties… Visualization Master headline

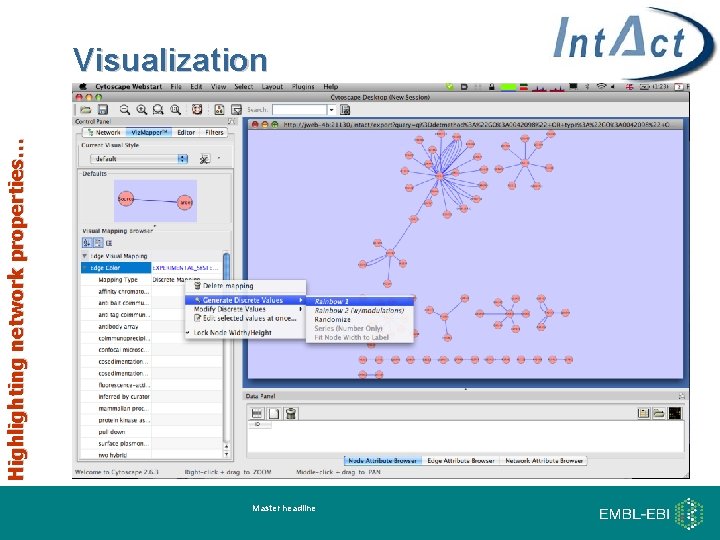
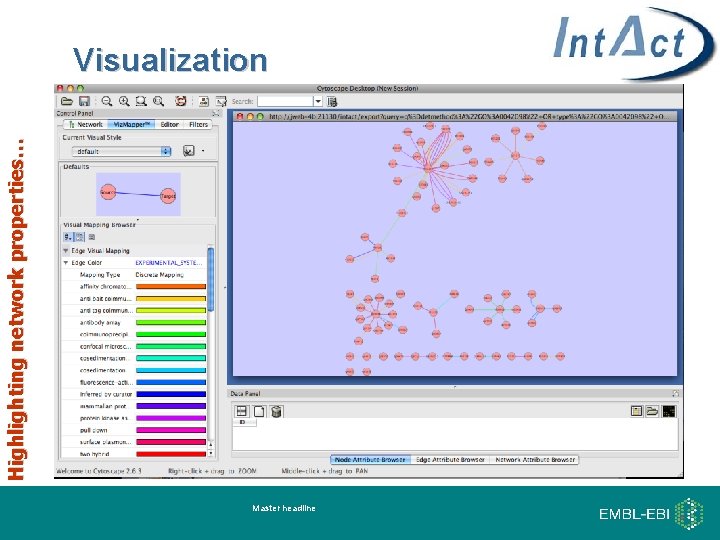
Highlighting network properties… Visualization Master headline

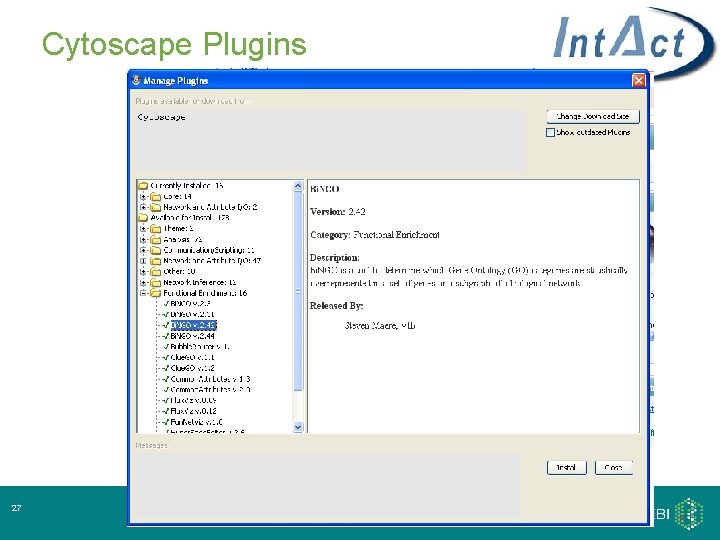
Cytoscape Plugins 27

Exploring a single interaction in more depth EBI is an Outstation of the European Molecular Biology Laboratory.

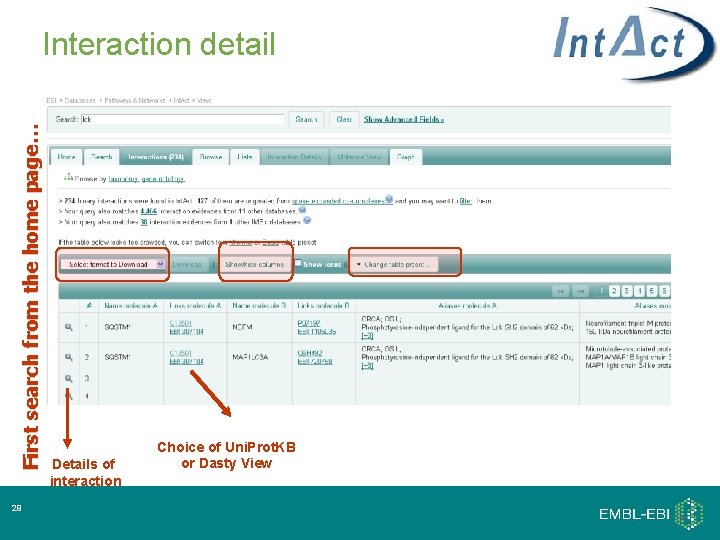
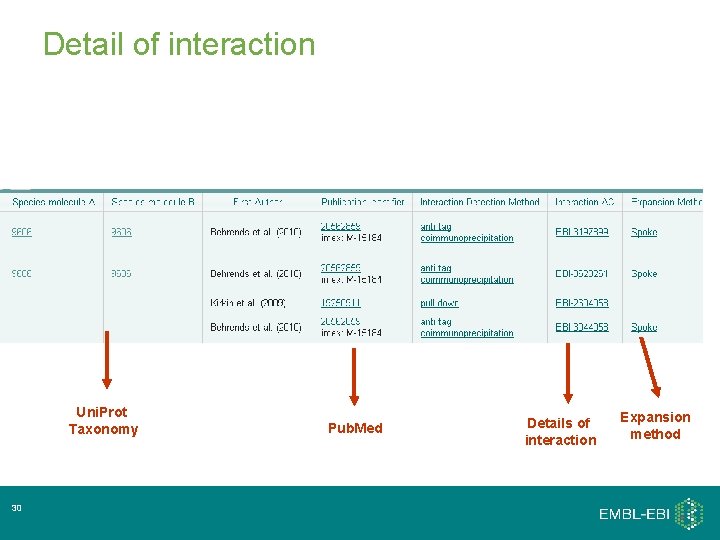
First search from the home page… Interaction detail 29 Details of interaction Choice of Uni. Prot. KB or Dasty View

Detail of interaction Uni. Prot Taxonomy 30 Pub. Med Details of interaction Expansion method

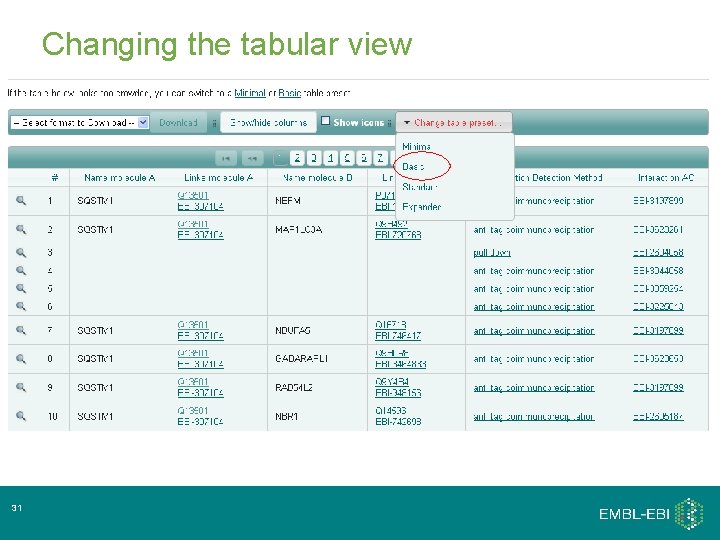
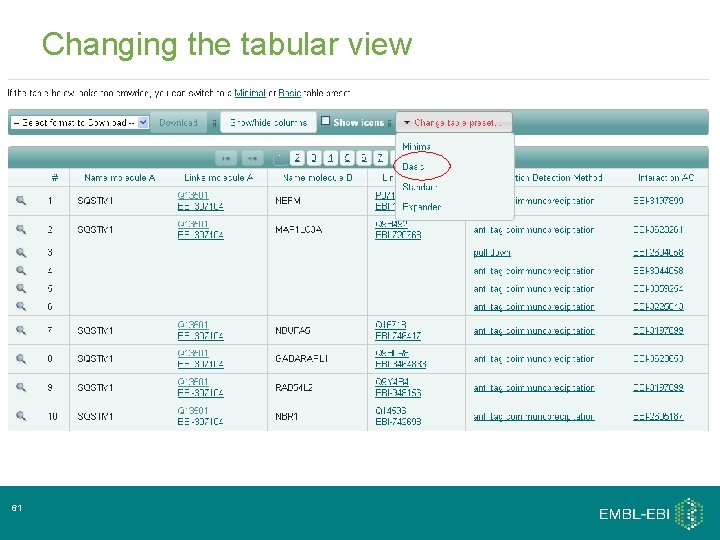
Changing the tabular view 31

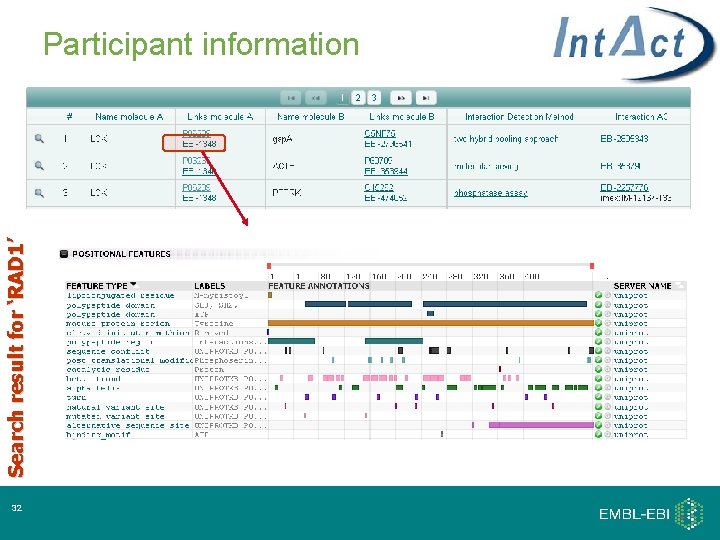
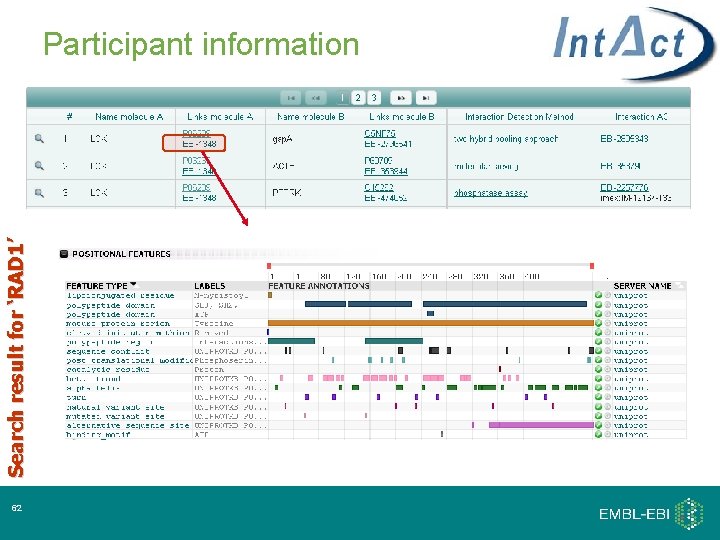
Search result for ‘RAD 1’ Participant information 32

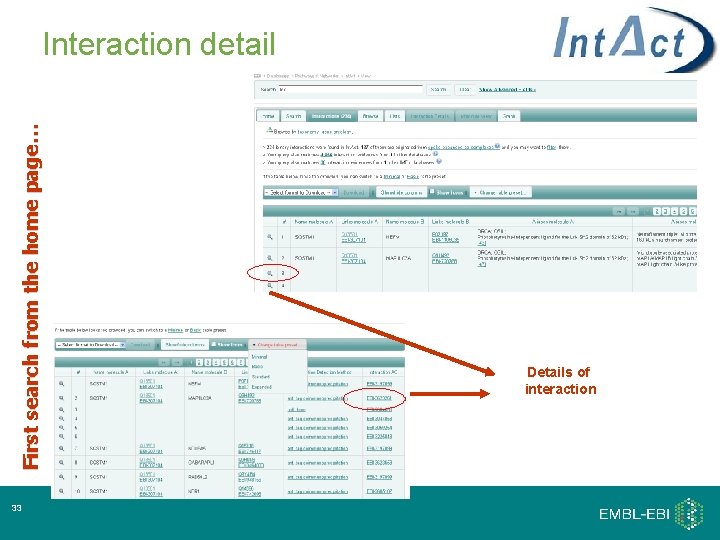
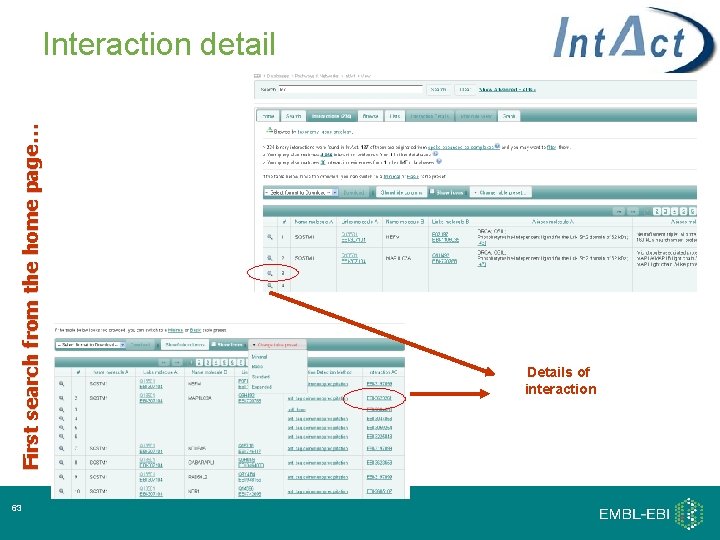
First search from the home page… Interaction detail 33 Details of interaction

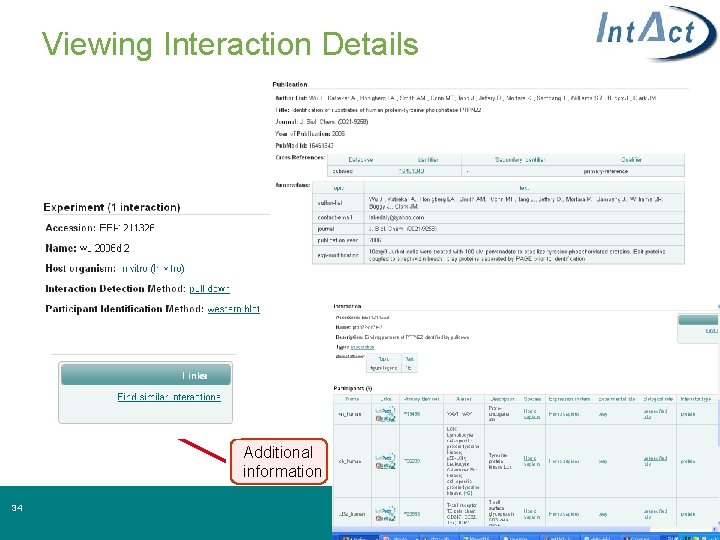
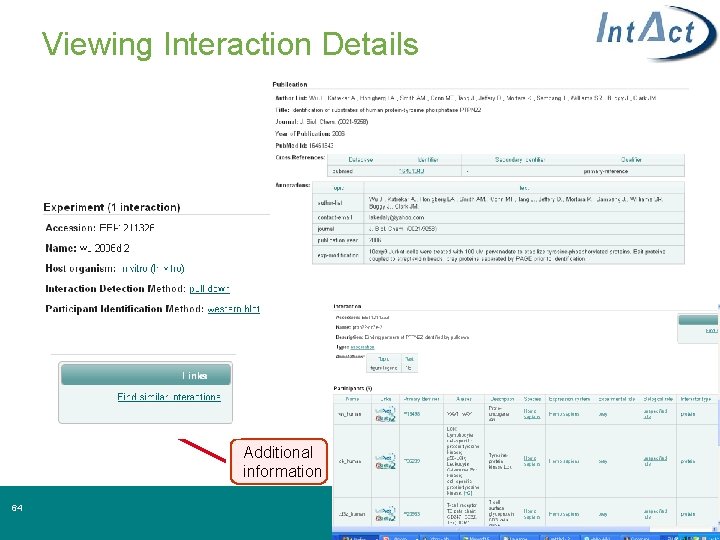
Viewing Interaction Details Additional information 34

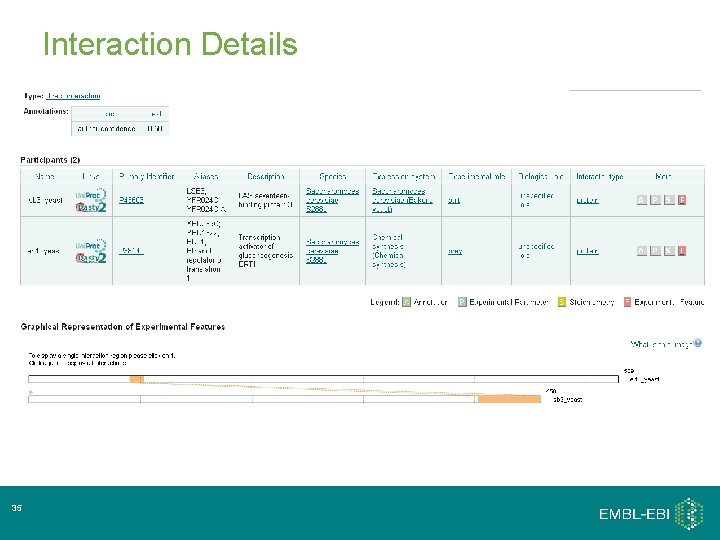
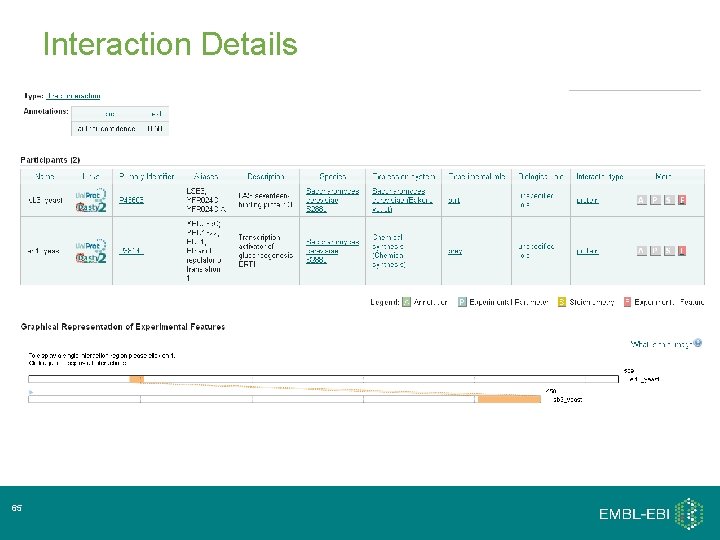
Interaction Details 35

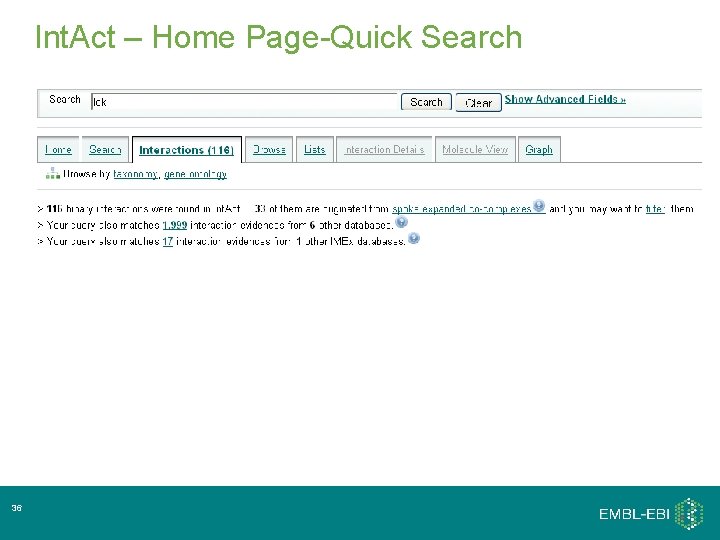
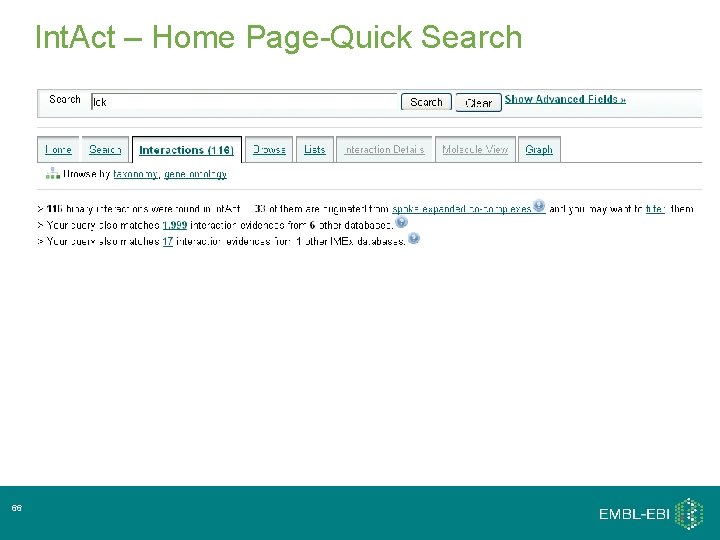
Int. Act – Home Page-Quick Search 36

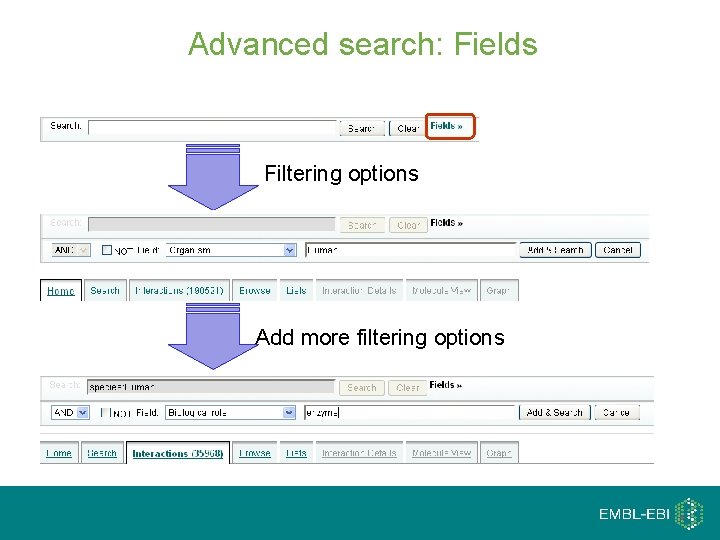
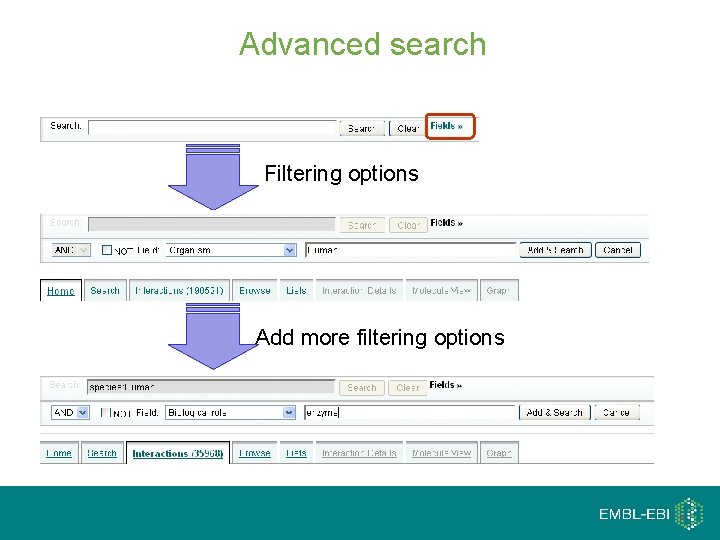
Advanced search: Fields Filtering options Add more filtering options

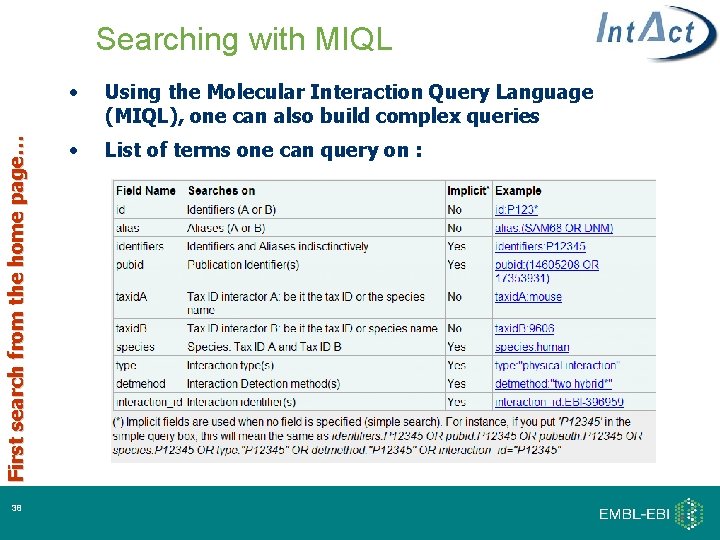
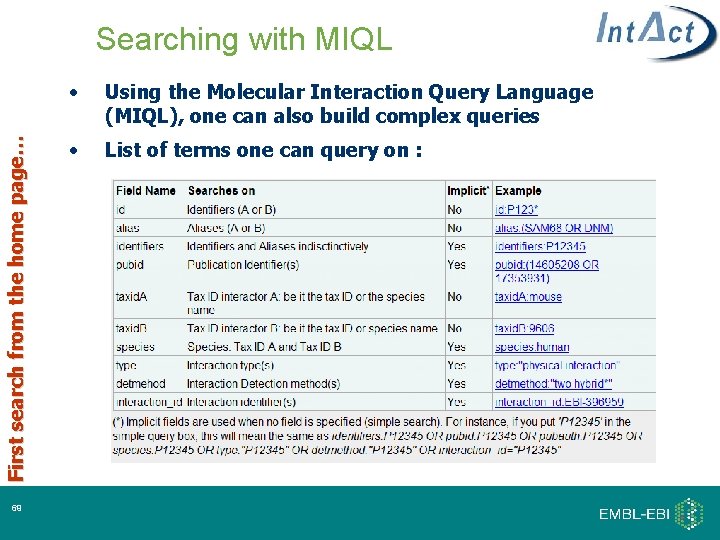
First search from the home page… Searching with MIQL 38 • Using the Molecular Interaction Query Language (MIQL), one can also build complex queries • List of terms one can query on :

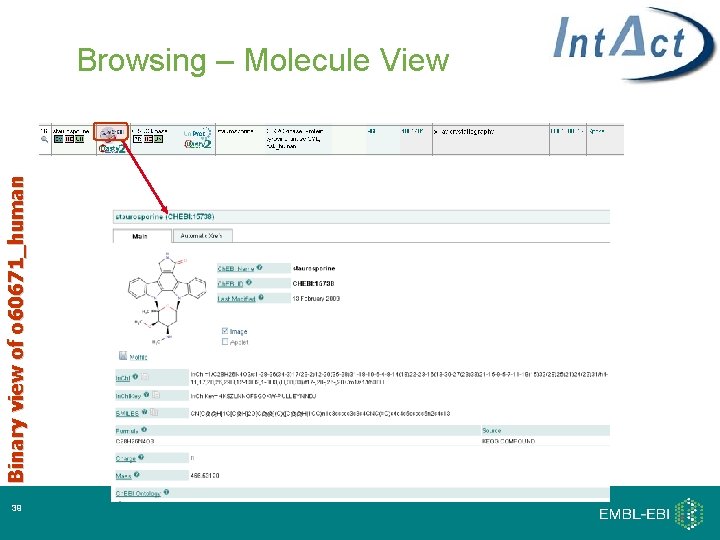
Binary view of o 60671_human Browsing – Molecule View 39

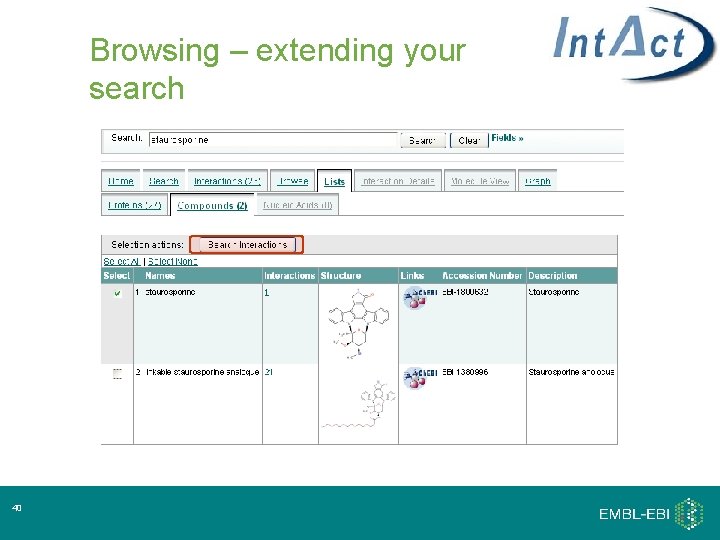
Browsing – extending your search 40


Performing and visualing a Simple Search Data, Standards and Tools EBI Walthrough May 2009 EBI is an Outstation of the European Molecular Biology Laboratory.

http: //www. ebi. ac. uk/intact Int. Act – Home Page 43

Performing a Simple Search 44

From search to network. View… Visualizing - network. View 45

Extend and Visualise your Search 46

Visualizing - network. View 47

Cytoscape Web • Cytoscape Web - web-based network visualization tool • Modeled after Cytoscape – open-source, interactive, customizable and easily integrated into web sites. • Contains none of the plugin architecture functionality of Cytoscape 48

Opening the network in Cytoscape… Visualization Master headline

Applying a better graph layout… Visualization Master headline

Applying a better graph layout… Visualization Master headline

Highlighting network properties… Visualization Master headline

Highlighting network properties… Visualization Master headline

Highlighting network properties… Visualization Master headline

Highlighting network properties… Visualization Master headline

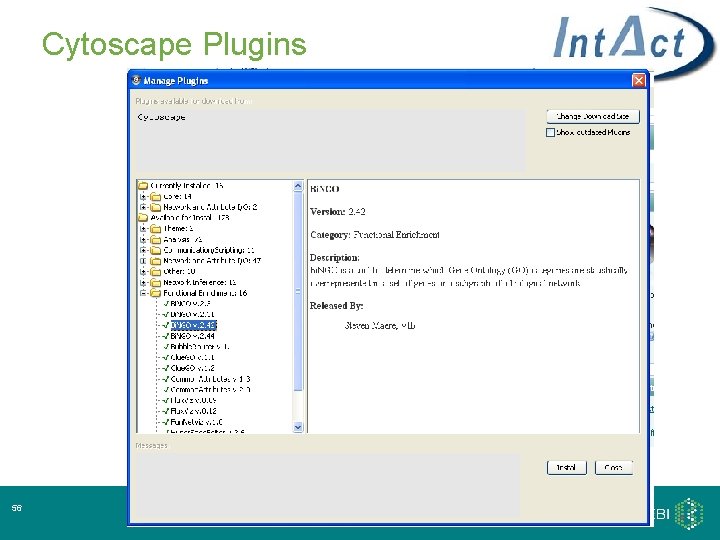
Cytoscape Plugins 56

Exploring a single interaction in more depth EBI is an Outstation of the European Molecular Biology Laboratory.

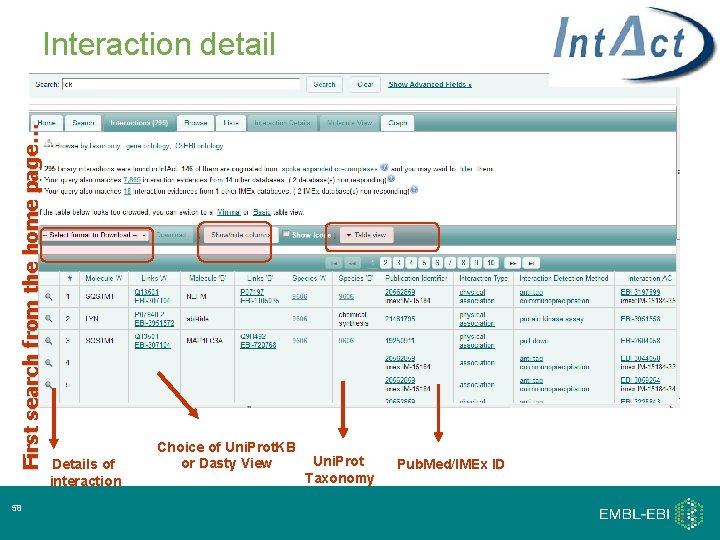
First search from the home page… Interaction detail 58 Details of interaction Choice of Uni. Prot. KB or Dasty View Uni. Prot Taxonomy Pub. Med/IMEx ID

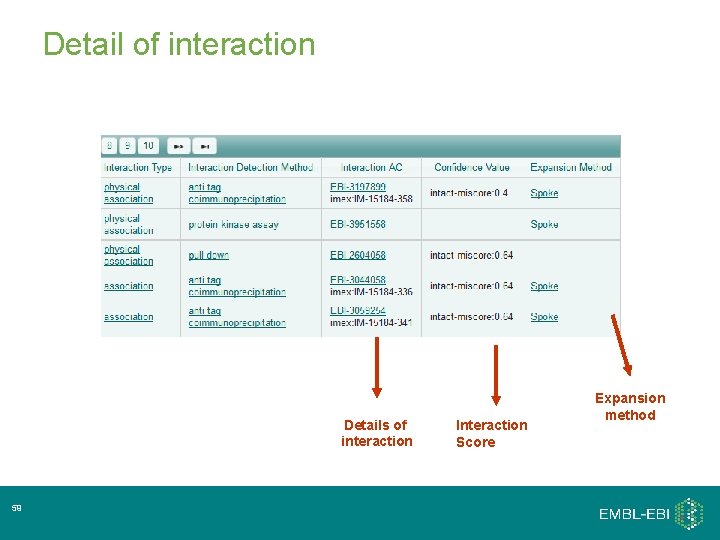
Detail of interaction Details of interaction 59 Interaction Score Expansion method

Interaction Score • All evidences of Protein A interacting with Protein B are clustered. • Evidences are scored according to a. Interaction detection method b. Interaction type c. Number of publications interaction has been observed in Score is normalised on 0 -1 scale Low score – low confidence interaction High score – high confidence interaction 60

Changing the tabular view 61

Search result for ‘RAD 1’ Participant information 62

First search from the home page… Interaction detail 63 Details of interaction

Viewing Interaction Details Additional information 64

Interaction Details 65

Int. Act – Home Page-Quick Search 66

Advanced search Filtering options Add more filtering options

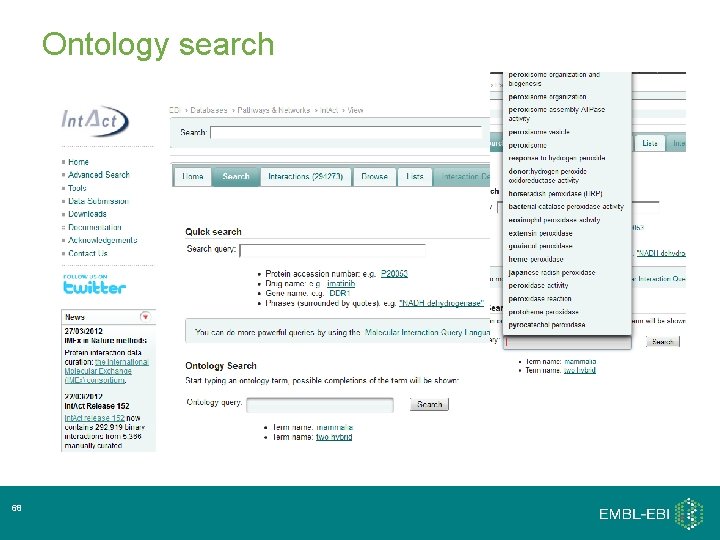
Ontology search 68

First search from the home page… Searching with MIQL 69 • Using the Molecular Interaction Query Language (MIQL), one can also build complex queries • List of terms one can query on :

Binary view of o 60671_human Browsing – Molecule View 70

Browsing – extending your search 71
