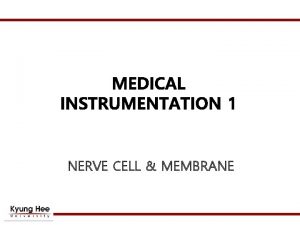
10 m Human height Length of some nerve








- Slides: 34

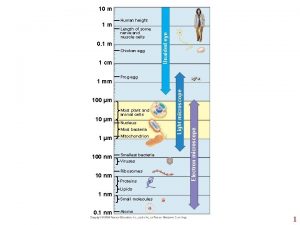
10 m Human height Length of some nerve and muscle cells 0. 1 m Chicken egg 1 cm 100 µm 10 µm Most plant and animal cells Nucleus Most bacteria 1 µm 100 nm 1 nm 0. 1 nm Mitochondrion Smallest bacteria Viruses Ribosomes Proteins Lipids Small molecules Atoms Electron microscope 1 mm Frog egg Light microscope 1 m Unaided eye Fig. 6 -2

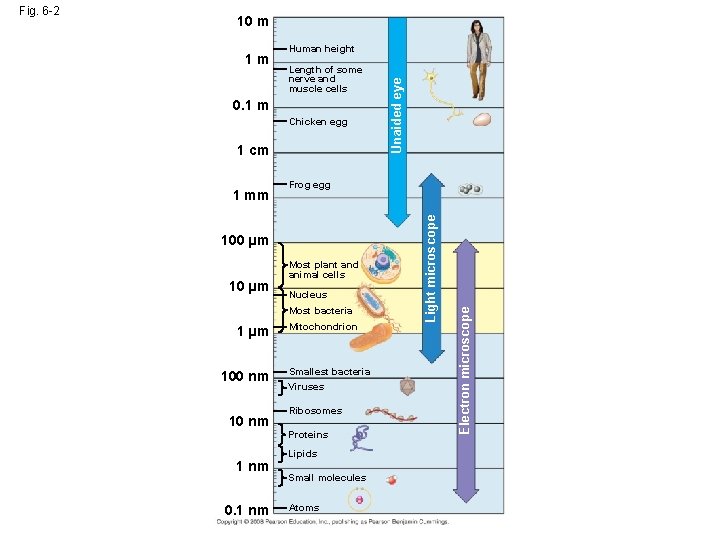
Fig. 6 -8 Surface area increases while total volume remains constant 5 1 1 Total surface area [Sum of the surface areas (height width) of all boxes sides number of boxes] Total volume [height width length number of boxes] Surface-to-volume (S-to-V) ratio [surface area ÷ volume] 6 150 750 1 125 6 1. 2 6

Fig. 6 -6 Fimbriae Nucleoid Ribosomes Plasma membrane Bacterial chromosome (a) A typical rod-shaped bacterium Cell wall Capsule Flagella 0. 5 µm (b) A thin section through the bacterium Bacillus coagulans (TEM)

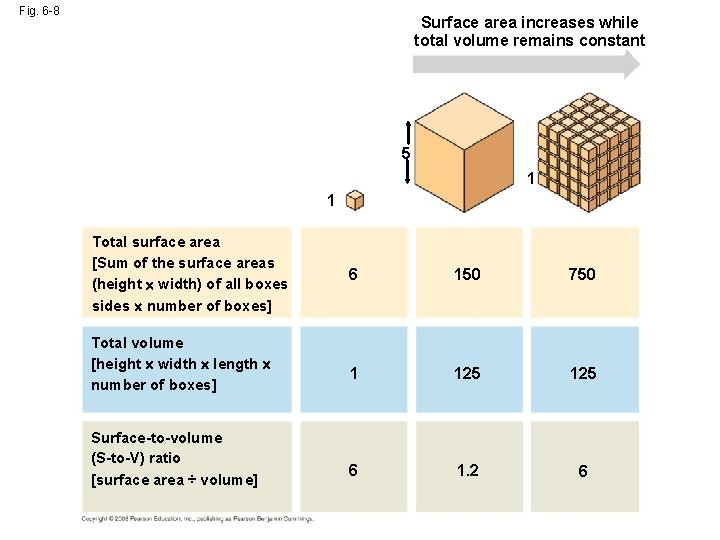
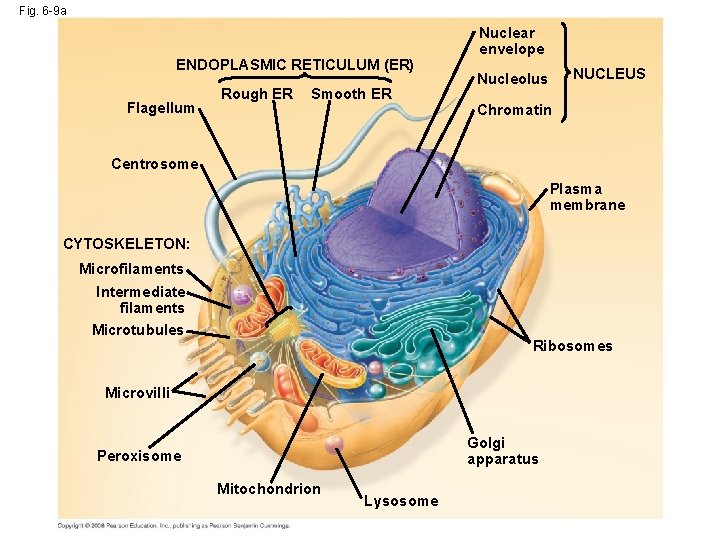
Fig. 6 -9 a ENDOPLASMIC RETICULUM (ER) Flagellum Rough ER Smooth ER Nuclear envelope NUCLEUS Nucleolus Chromatin Centrosome Plasma membrane CYTOSKELETON: Microfilaments Intermediate filaments Microtubules Ribosomes Microvilli Golgi apparatus Peroxisome Mitochondrion Lysosome

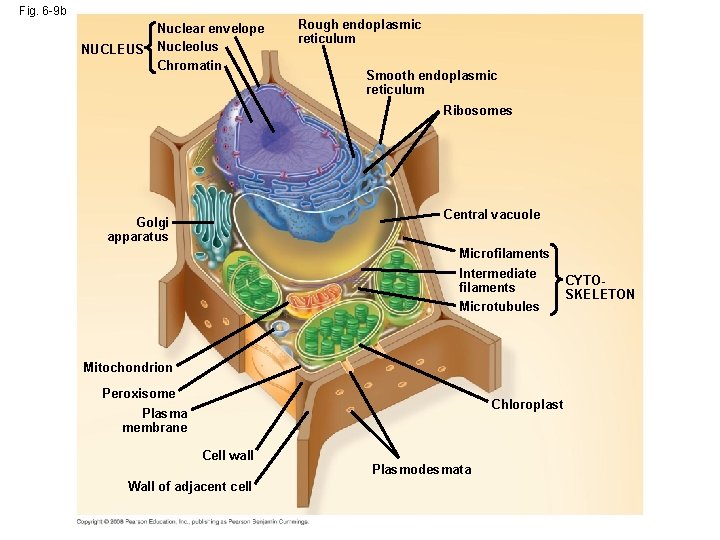
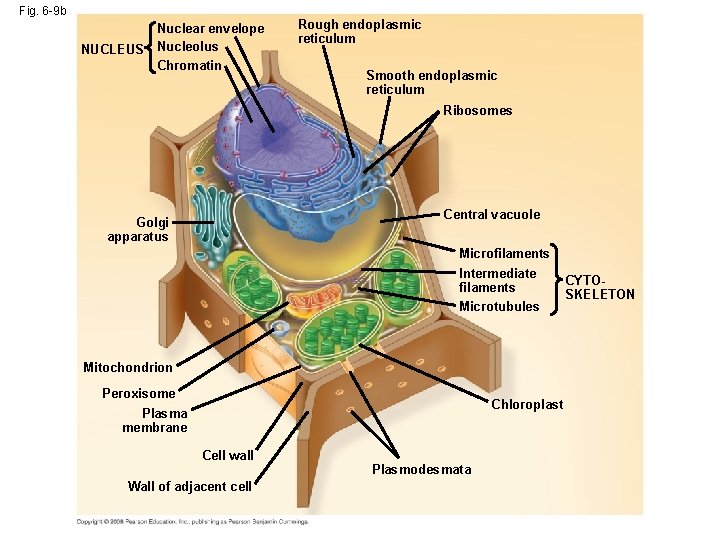
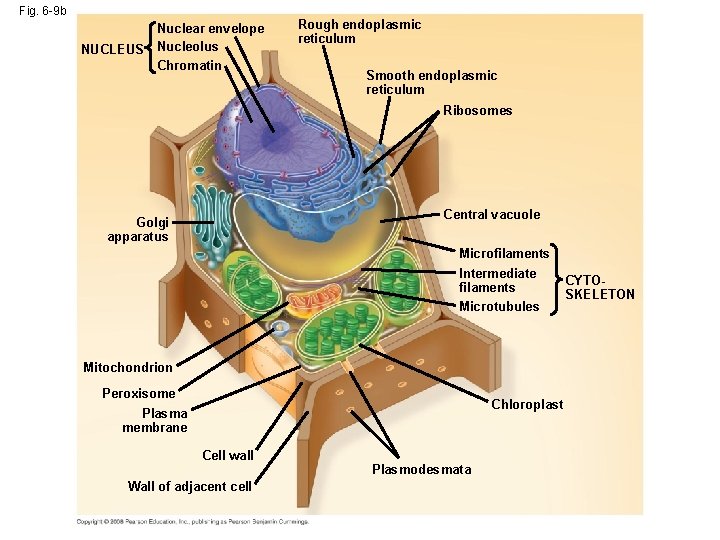
Fig. 6 -9 b NUCLEUS Nuclear envelope Nucleolus Chromatin Rough endoplasmic reticulum Smooth endoplasmic reticulum Ribosomes Central vacuole Golgi apparatus Microfilaments Intermediate filaments Microtubules Mitochondrion Peroxisome Chloroplast Plasma membrane Cell wall Wall of adjacent cell Plasmodesmata CYTOSKELETON

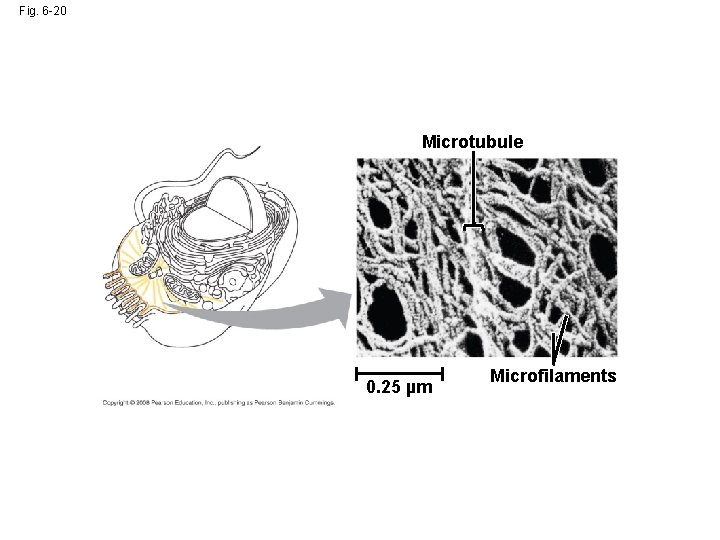
Fig. 6 -20 Microtubule 0. 25 µm Microfilaments

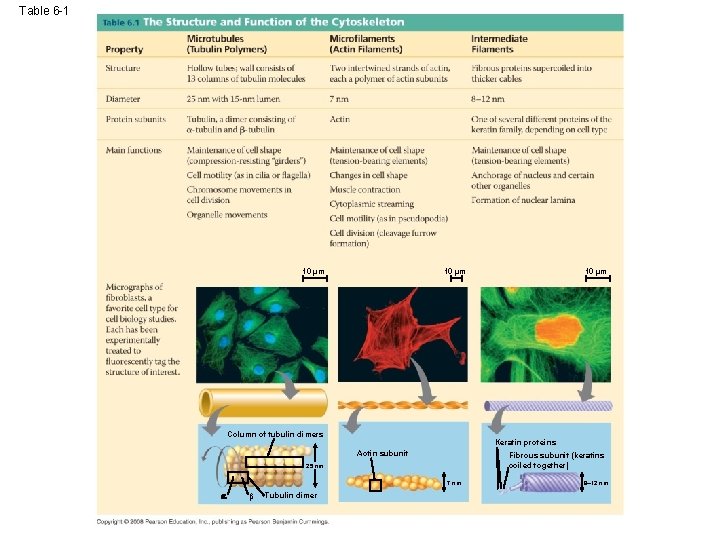
Table 6 -1 10 µm Column of tubulin dimers Keratin proteins Actin subunit Fibrous subunit (keratins coiled together) 25 nm 7 nm Tubulin dimer 10 µm 8– 12 nm

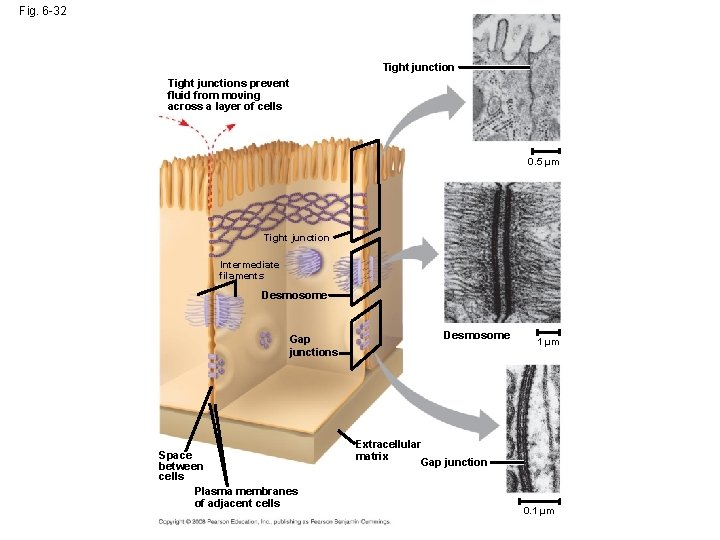
Fig. 6 -32 Tight junctions prevent fluid from moving across a layer of cells 0. 5 µm Tight junction Intermediate filaments Desmosome Gap junctions Space between cells Plasma membranes of adjacent cells Desmosome 1 µm Extracellular matrix Gap junction 0. 1 µm

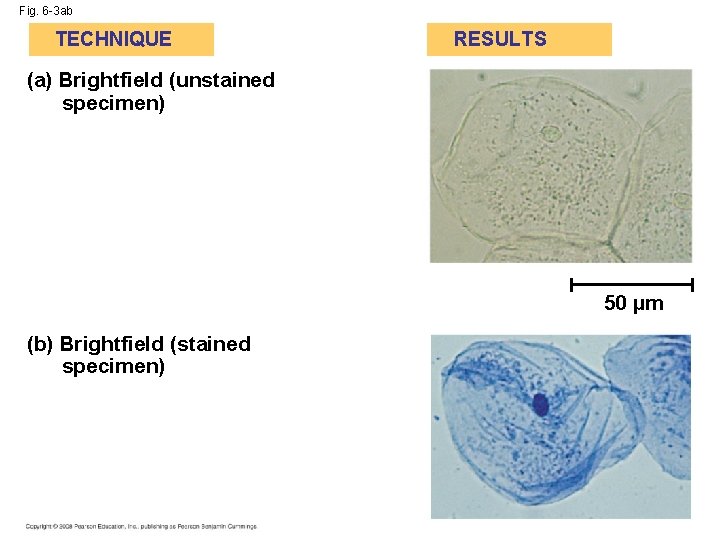
Fig. 6 -3 ab TECHNIQUE RESULTS (a) Brightfield (unstained specimen) 50 µm (b) Brightfield (stained specimen)

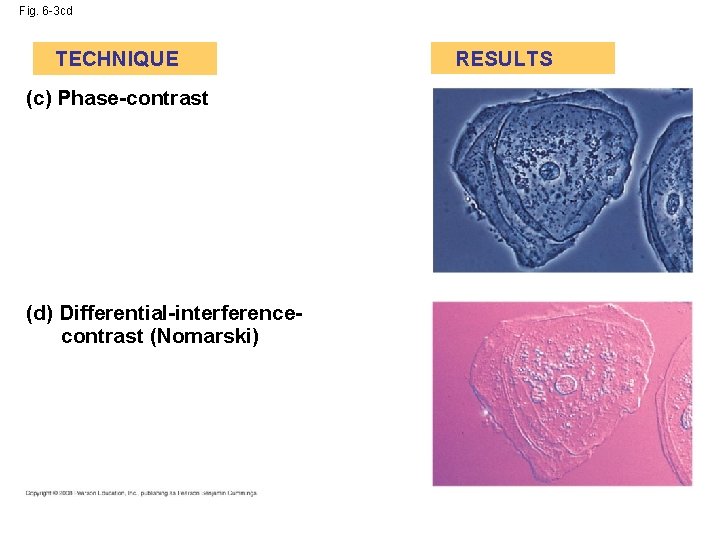
Fig. 6 -3 cd TECHNIQUE (c) Phase-contrast (d) Differential-interferencecontrast (Nomarski) RESULTS

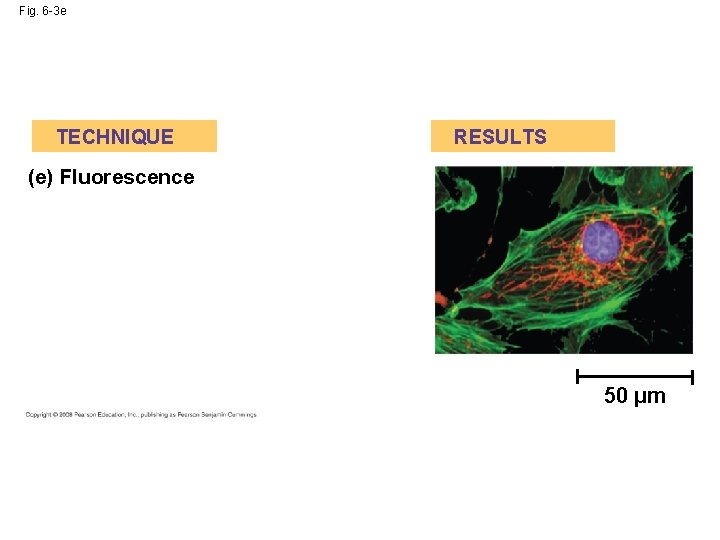
Fig. 6 -3 e TECHNIQUE RESULTS (e) Fluorescence 50 µm

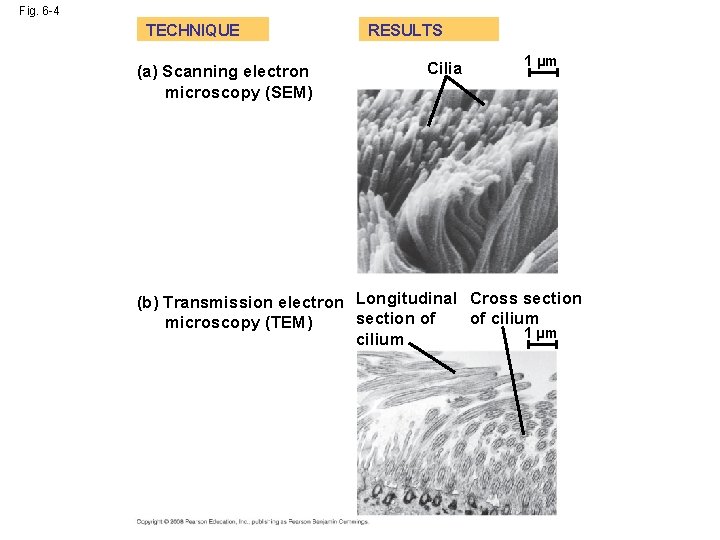
Fig. 6 -4 TECHNIQUE (a) Scanning electron microscopy (SEM) RESULTS Cilia 1 µm (b) Transmission electron Longitudinal Cross section of of cilium microscopy (TEM) 1 µm cilium

Fig. 6 -9 a ENDOPLASMIC RETICULUM (ER) Flagellum Rough ER Smooth ER Nuclear envelope NUCLEUS Nucleolus Chromatin Centrosome Plasma membrane CYTOSKELETON: Microfilaments Intermediate filaments Microtubules Ribosomes Microvilli Golgi apparatus Peroxisome Mitochondrion Lysosome

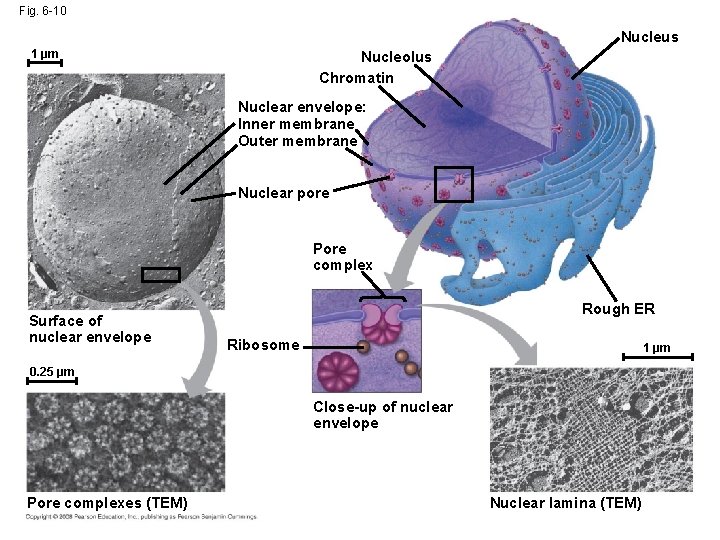
Fig. 6 -10 Nucleus 1 µm Nucleolus Chromatin Nuclear envelope: Inner membrane Outer membrane Nuclear pore Pore complex Surface of nuclear envelope Rough ER Ribosome 1 µm 0. 25 µm Close-up of nuclear envelope Pore complexes (TEM) Nuclear lamina (TEM)

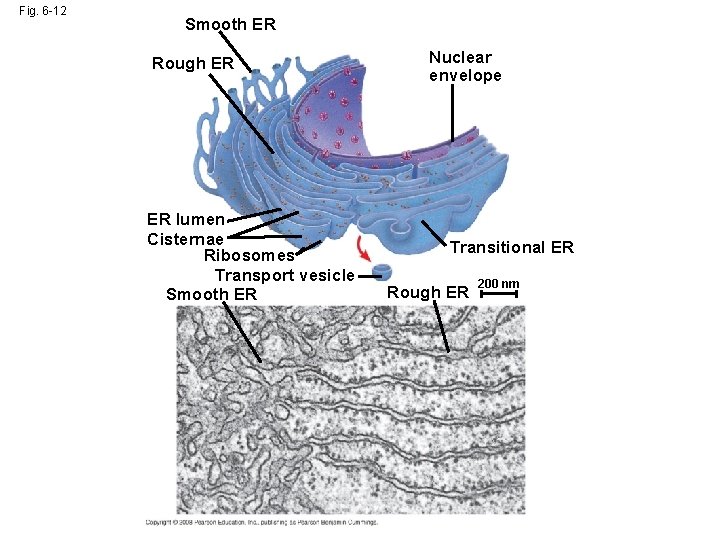
Fig. 6 -12 Smooth ER Rough ER ER lumen Cisternae Ribosomes Transport vesicle Smooth ER Nuclear envelope Transitional ER Rough ER 200 nm

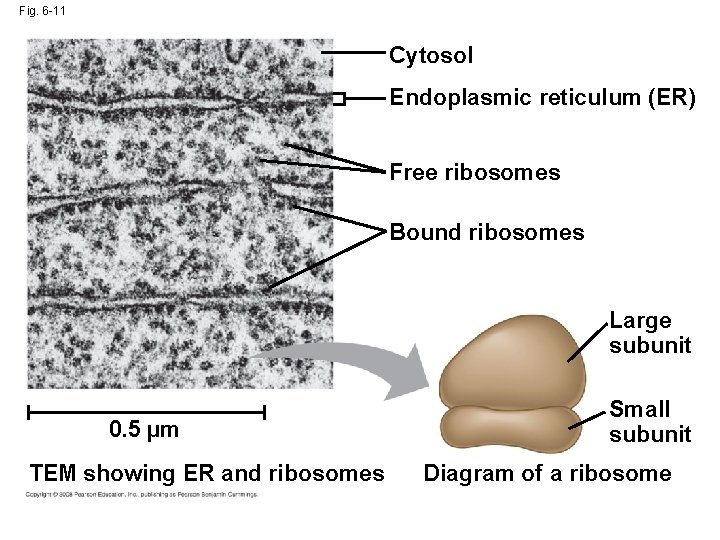
Fig. 6 -11 Cytosol Endoplasmic reticulum (ER) Free ribosomes Bound ribosomes Large subunit 0. 5 µm TEM showing ER and ribosomes Small subunit Diagram of a ribosome

Fig. 6 -12 Smooth ER Rough ER ER lumen Cisternae Ribosomes Transport vesicle Smooth ER Nuclear envelope Transitional ER Rough ER 200 nm

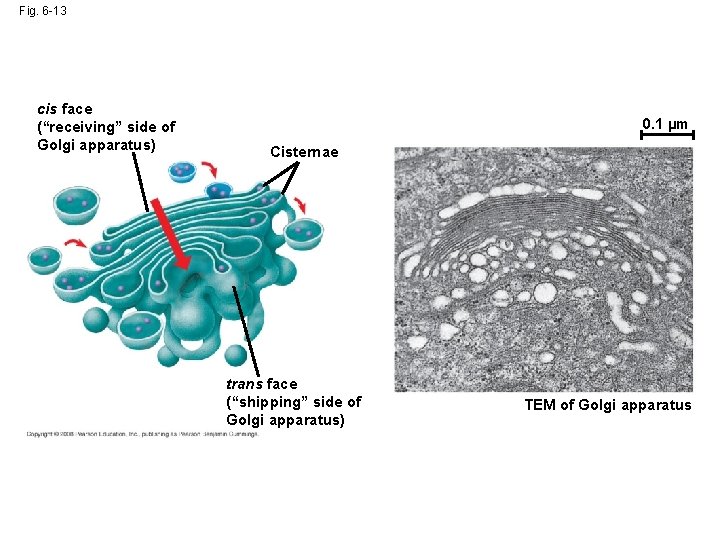
Fig. 6 -13 cis face (“receiving” side of Golgi apparatus) 0. 1 µm Cisternae trans face (“shipping” side of Golgi apparatus) TEM of Golgi apparatus

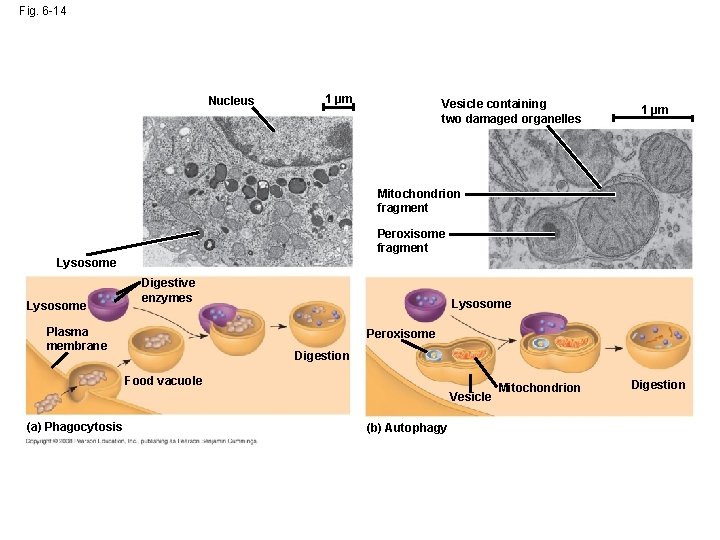
Fig. 6 -14 Nucleus 1 µm Vesicle containing two damaged organelles 1 µm Mitochondrion fragment Peroxisome fragment Lysosome Digestive enzymes Plasma membrane Lysosome Peroxisome Digestion Food vacuole Vesicle (a) Phagocytosis (b) Autophagy Mitochondrion Digestion

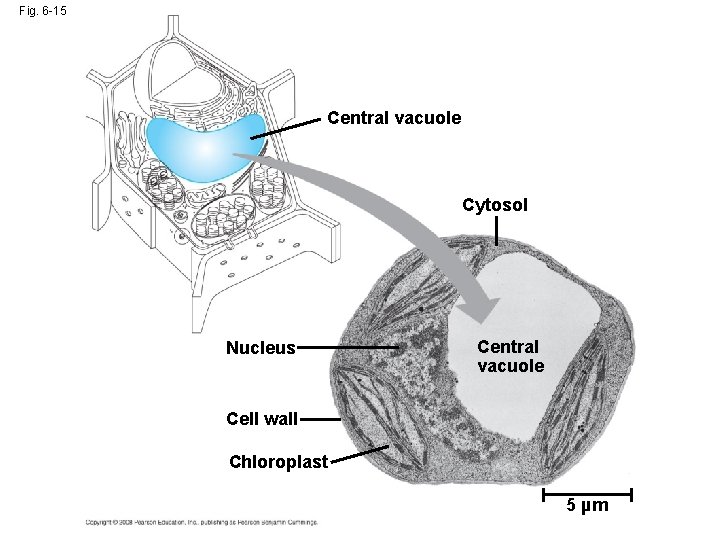
Fig. 6 -15 Central vacuole Cytosol Nucleus Central vacuole Cell wall Chloroplast 5 µm

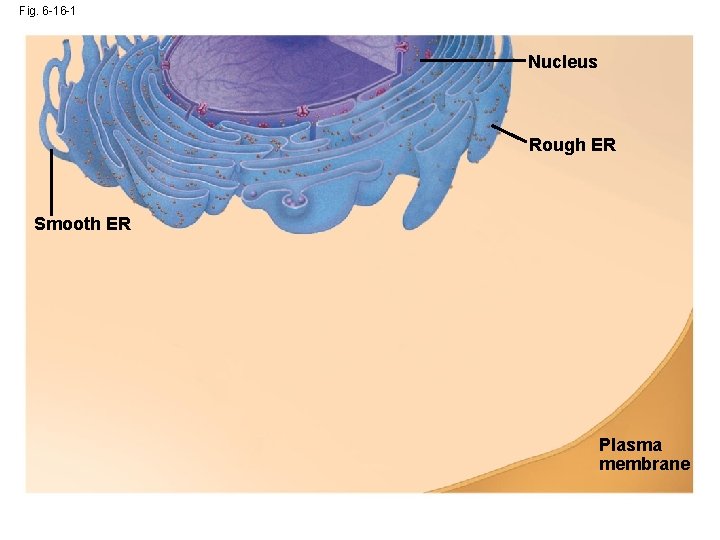
Fig. 6 -16 -1 Nucleus Rough ER Smooth ER Plasma membrane

Fig. 6 -16 -2 Nucleus Rough ER Smooth ER cis Golgi trans Golgi Plasma membrane

Fig. 6 -16 -3 Nucleus Rough ER Smooth ER cis Golgi trans Golgi Plasma membrane

Fig. 6 -14 Nucleus 1 µm Vesicle containing two damaged organelles 1 µm Mitochondrion fragment Peroxisome fragment Lysosome Digestive enzymes Plasma membrane Lysosome Peroxisome Digestion Food vacuole Vesicle (a) Phagocytosis (b) Autophagy Mitochondrion Digestion

Fig. 6 -9 b NUCLEUS Nuclear envelope Nucleolus Chromatin Rough endoplasmic reticulum Smooth endoplasmic reticulum Ribosomes Central vacuole Golgi apparatus Microfilaments Intermediate filaments Microtubules Mitochondrion Peroxisome Chloroplast Plasma membrane Cell wall Wall of adjacent cell Plasmodesmata CYTOSKELETON

Fig. 6 -9 a ENDOPLASMIC RETICULUM (ER) Flagellum Rough ER Smooth ER Nuclear envelope NUCLEUS Nucleolus Chromatin Centrosome Plasma membrane CYTOSKELETON: Microfilaments Intermediate filaments Microtubules Ribosomes Microvilli Golgi apparatus Peroxisome Mitochondrion Lysosome

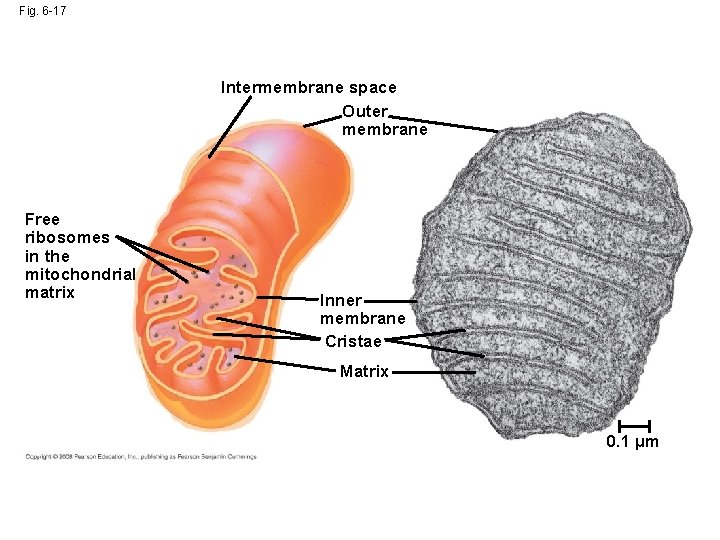
Fig. 6 -17 Intermembrane space Outer membrane Free ribosomes in the mitochondrial matrix Inner membrane Cristae Matrix 0. 1 µm

Fig. 6 -9 b NUCLEUS Nuclear envelope Nucleolus Chromatin Rough endoplasmic reticulum Smooth endoplasmic reticulum Ribosomes Central vacuole Golgi apparatus Microfilaments Intermediate filaments Microtubules Mitochondrion Peroxisome Chloroplast Plasma membrane Cell wall Wall of adjacent cell Plasmodesmata CYTOSKELETON

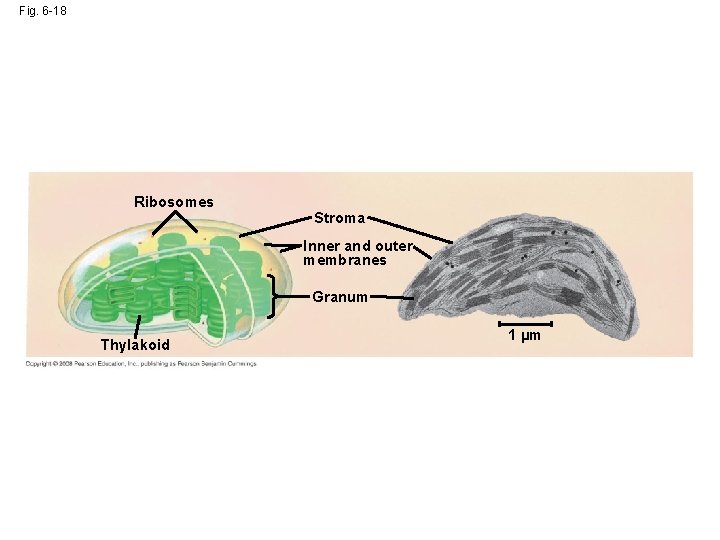
Fig. 6 -18 Ribosomes Stroma Inner and outer membranes Granum Thylakoid 1 µm

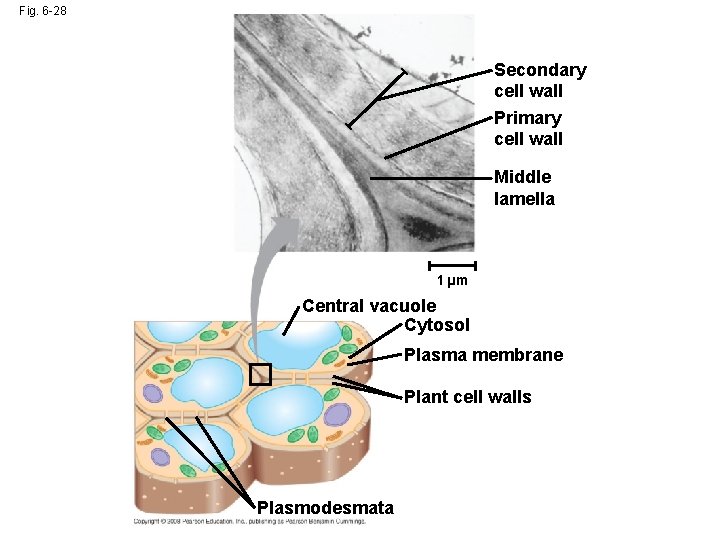
Fig. 6 -28 Secondary cell wall Primary cell wall Middle lamella 1 µm Central vacuole Cytosol Plasma membrane Plant cell walls Plasmodesmata

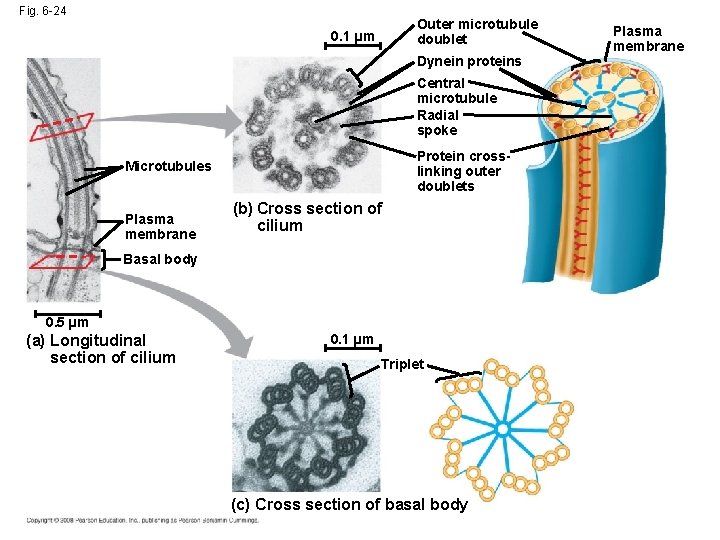
Fig. 6 -24 Outer microtubule doublet 0. 1 µm Dynein proteins Central microtubule Radial spoke Protein crosslinking outer doublets Microtubules Plasma membrane (b) Cross section of cilium Basal body 0. 5 µm (a) Longitudinal section of cilium 0. 1 µm Triplet (c) Cross section of basal body Plasma membrane

Fig. 6 -25 Microtubule doublets ATP Dynein protein (a) Effect of unrestrained dynein movement ATP Cross-linking proteins inside outer doublets Anchorage in cell (b) Effect of cross-linking proteins 1 3 2 (c) Wavelike motion

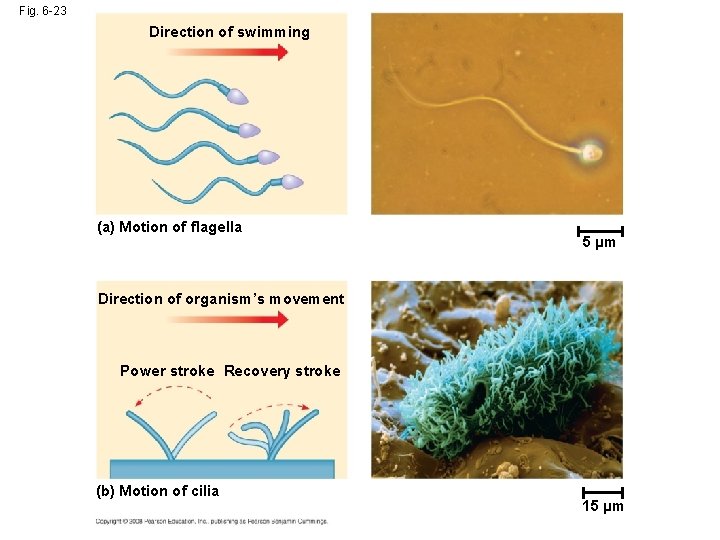
Fig. 6 -23 Direction of swimming (a) Motion of flagella 5 µm Direction of organism’s movement Power stroke Recovery stroke (b) Motion of cilia 15 µm

Fig. 6 -22 Centrosome Microtubule Centrioles 0. 25 µm Longitudinal section Microtubules Cross section of one centriole of the other centriole
 Lc gate height gauge
Lc gate height gauge Length width and height of a rectangular prism
Length width and height of a rectangular prism Surface area of a parallelogram
Surface area of a parallelogram A=lxw example
A=lxw example The mass or general outline of a hairstyle
The mass or general outline of a hairstyle What are the 5 principles of hair design
What are the 5 principles of hair design H 319
H 319 Volume * density = mass
Volume * density = mass Determining height from bone length
Determining height from bone length What is the ratio of the length of to the length of ?
What is the ratio of the length of to the length of ? Length of optic nerve
Length of optic nerve Trigeminal nerve which cranial nerve
Trigeminal nerve which cranial nerve They say sometimes you win some
They say sometimes you win some Sometimes you win some sometimes you lose some
Sometimes you win some sometimes you lose some Ice cream count or noncount noun
Ice cream count or noncount noun Contact vs noncontact forces
Contact vs noncontact forces Some say the world will end in fire some say in ice
Some say the world will end in fire some say in ice Some say the world will end in fire some say in ice
Some say the world will end in fire some say in ice Some trust in horses
Some trust in horses Human height normal distribution
Human height normal distribution Hyoglossus muscle
Hyoglossus muscle Key issue 4 why are some human actions not sustainable
Key issue 4 why are some human actions not sustainable Human vs non human bones
Human vs non human bones Cna chapter 8 human needs and human development
Cna chapter 8 human needs and human development Chapter 8 human needs and human development
Chapter 8 human needs and human development Human development index definition ap human geography
Human development index definition ap human geography Non human nouns
Non human nouns What are votive figures
What are votive figures Height of cylinder gas
Height of cylinder gas Diameter of breast height
Diameter of breast height Abe hum pen thin height meaning
Abe hum pen thin height meaning Base x height example
Base x height example Volleyball
Volleyball Vision4you
Vision4you Gearless elevator
Gearless elevator