1 2 Ester functional group acid alcohol Handout

![6 Phospholipids: [HO] Handout 2 -9 6 Phospholipids: [HO] Handout 2 -9](https://slidetodoc.com/presentation_image/3e1aab3d1f0464702aa6fd9d15a2ee3f/image-6.jpg)






- Slides: 48

1

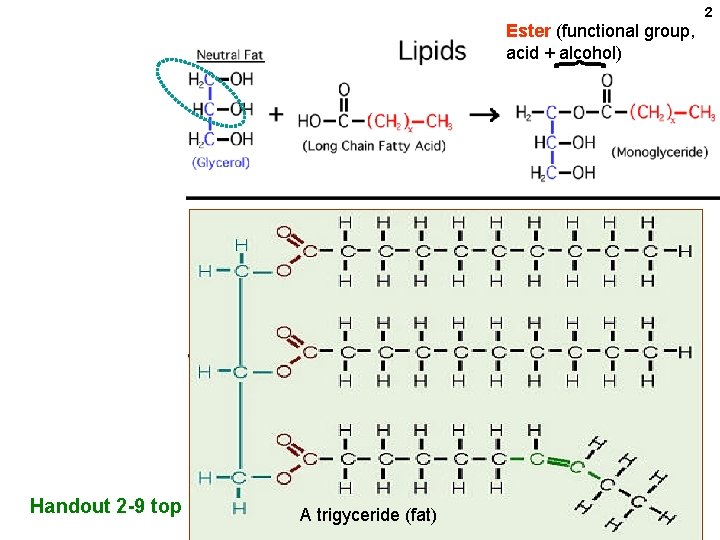
2 Ester (functional group, acid + alcohol) Handout 2 -9 top A trigyceride (fat)

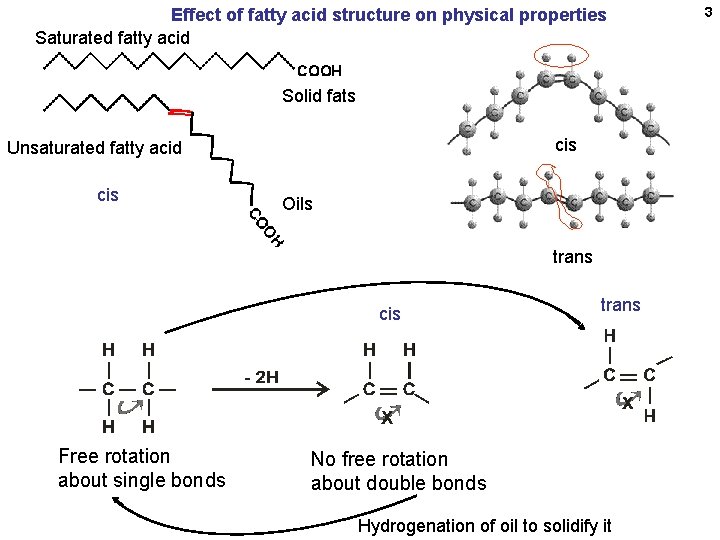
Effect of fatty acid structure on physical properties Saturated fatty acid Solid fats cis Unsaturated fatty acid cis Oils trans cis Free rotation about single bonds trans No free rotation about double bonds Hydrogenation of oil to solidify it 3

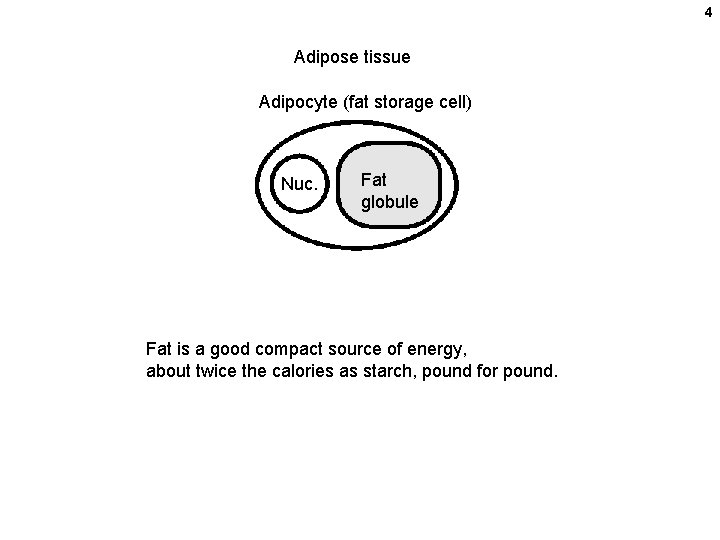
4 Adipose tissue Adipocyte (fat storage cell) Nuc. Fat globule Fat is a good compact source of energy, about twice the calories as starch, pound for pound.

5
![6 Phospholipids HO Handout 2 9 6 Phospholipids: [HO] Handout 2 -9](https://slidetodoc.com/presentation_image/3e1aab3d1f0464702aa6fd9d15a2ee3f/image-6.jpg)
6 Phospholipids: [HO] Handout 2 -9

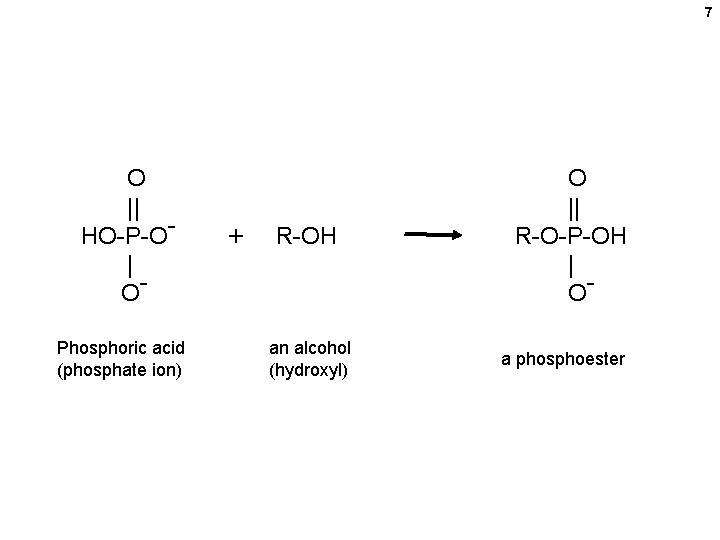
7 O || HO-P-O| OPhosphoric acid (phosphate ion) + R-OH an alcohol (hydroxyl) O || R-O-P-OH | Oa phosphoester

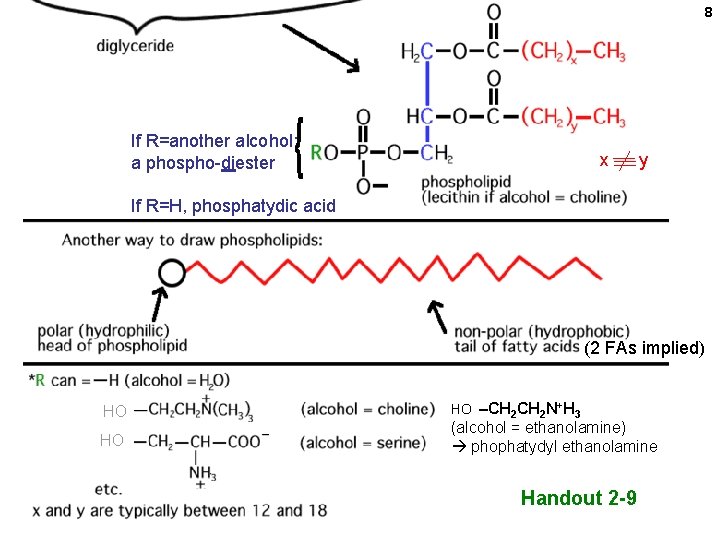
8 If R=another alcohol: a phospho-diester x y If R=H, phosphatydic acid (2 FAs implied) HO HO HO –CH 2 N+H 3 (alcohol = ethanolamine) phophatydyl ethanolamine Handout 2 -9

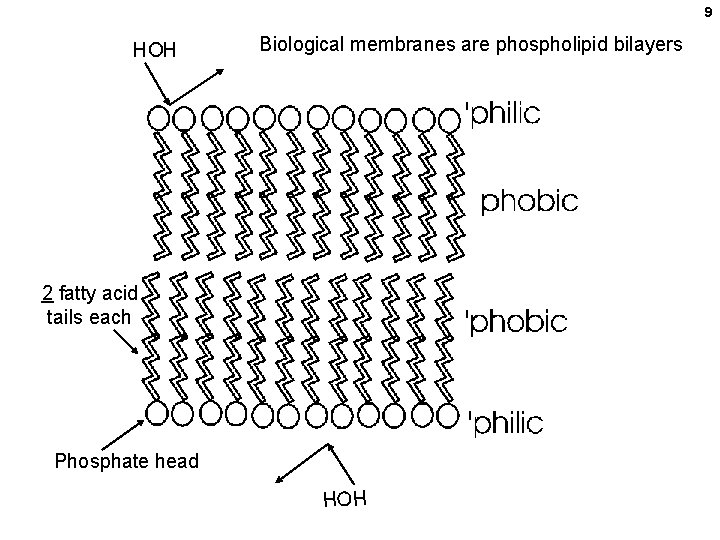
9 HOH Biological membranes are phospholipid bilayers 2 fatty acid tails each Phosphate head HOH

10 Incidentally, note the functional groups we have met so far: Hydroxyl Amine Amide Carboxyl Carbonyl Aldehyde Ketone Ester: Carboxylic acid ester Phosphodiester And: Glycosidic bonds C=C double bonds (cis and trans)

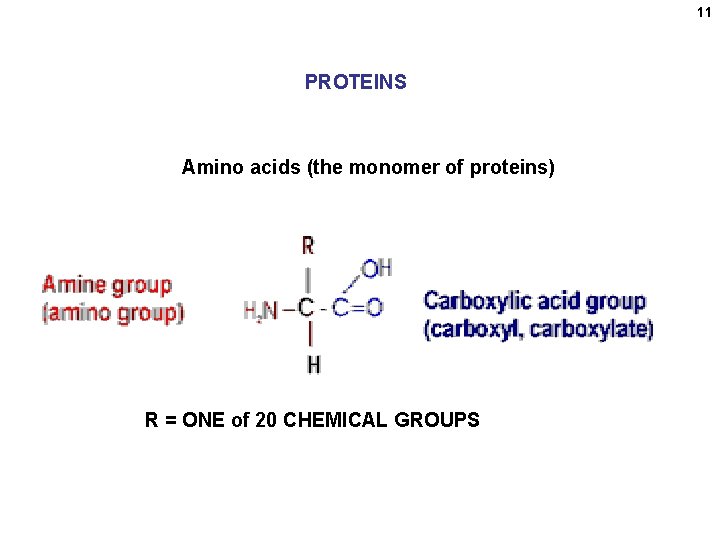
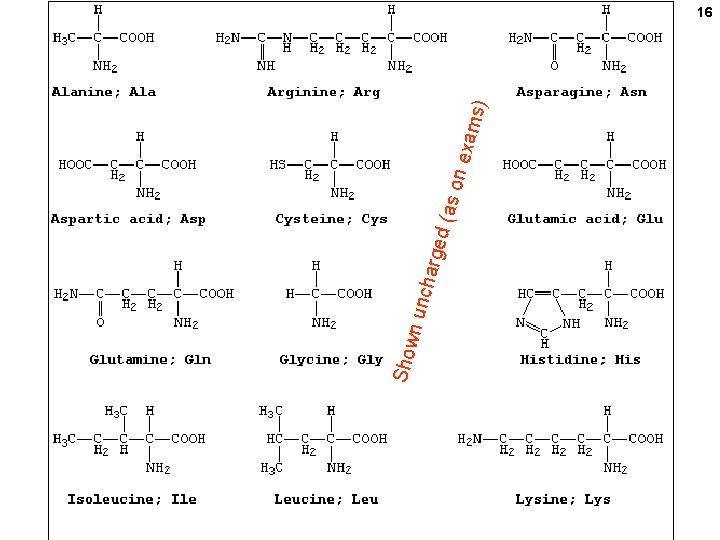
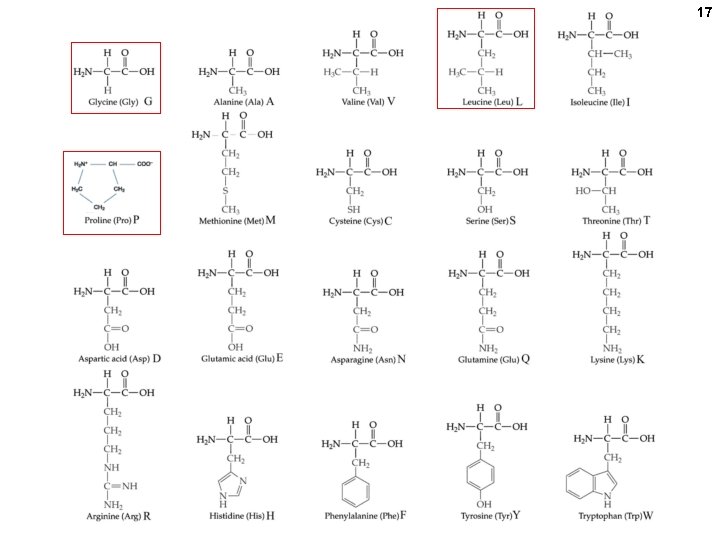
11 PROTEINS Amino acids (the monomer of proteins) R = ONE of 20 CHEMICAL GROUPS

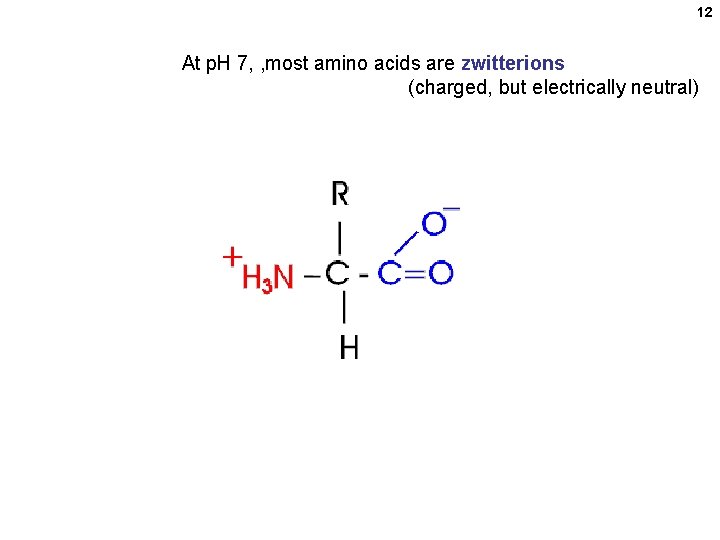
12 At p. H 7, , most amino acids are zwitterions (charged, but electrically neutral)

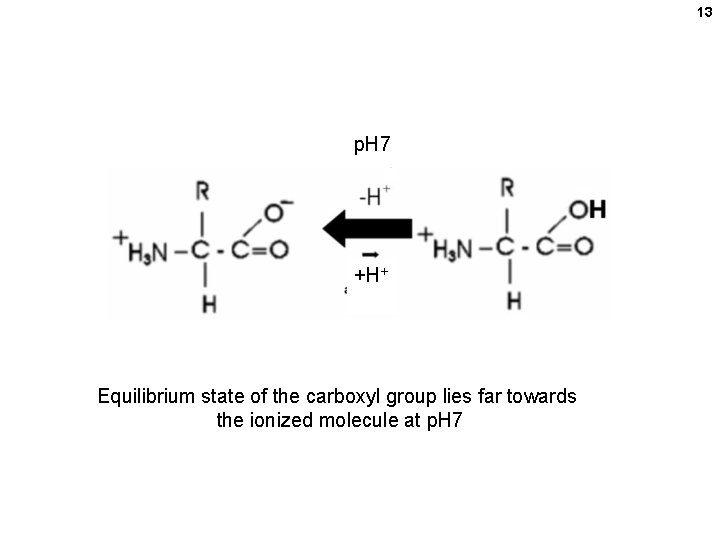
13 p. H 7 +H+ Equilibrium state of the carboxyl group lies far towards the ionized molecule at p. H 7

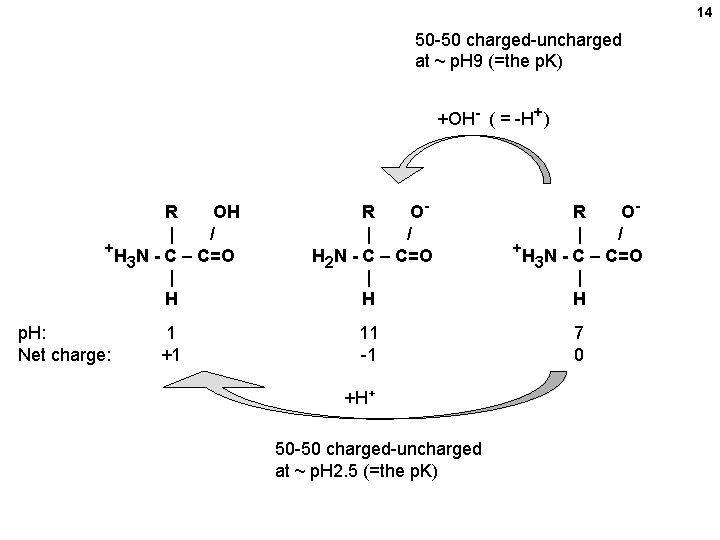
14 50 -50 charged-uncharged at ~ p. H 9 (=the p. K) +OH- ( = -H+) R OH | / +H N - C – C=O 3 | H p. H: Net charge: 1 +1 R O| / H 2 N - C – C=O | H R O| / +H N - C – C=O 3 | H 11 -1 7 0 +H+ 50 -50 charged-uncharged at ~ p. H 2. 5 (=the p. K)

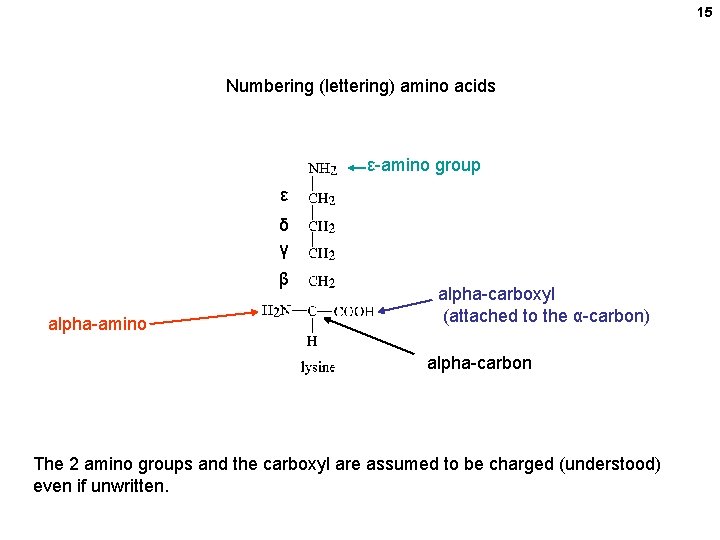
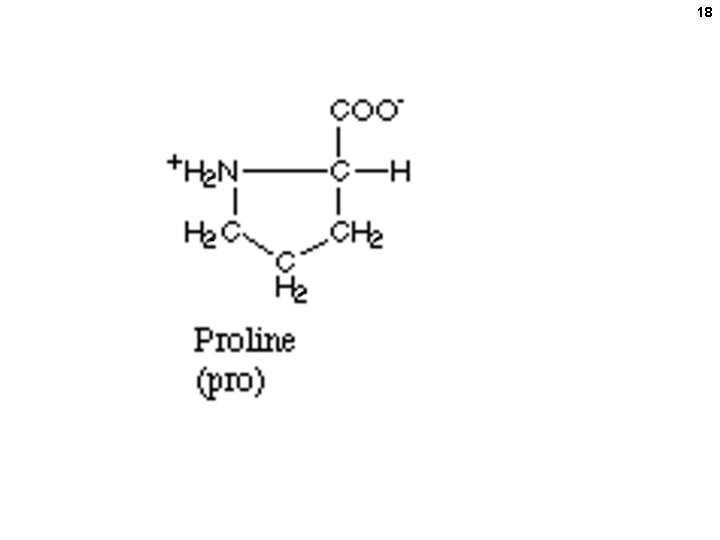
15 Numbering (lettering) amino acids ε-amino group ε δ γ β alpha-amino alpha-carboxyl (attached to the α-carbon) alpha-carbon The 2 amino groups and the carboxyl are assumed to be charged (understood) even if unwritten.

Show ged (a char n un s on exam s) 16

17

18

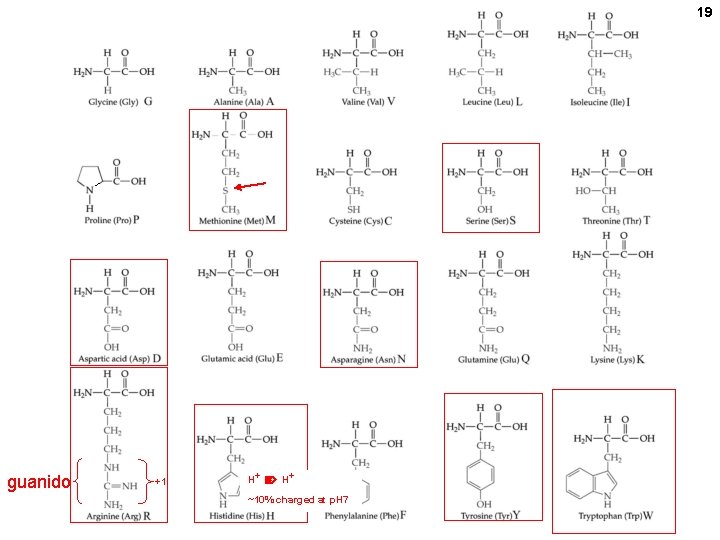
19 guanido +1 H+ ~10% charged at p. H 7

20 Ball and stick physical model of an amino acid

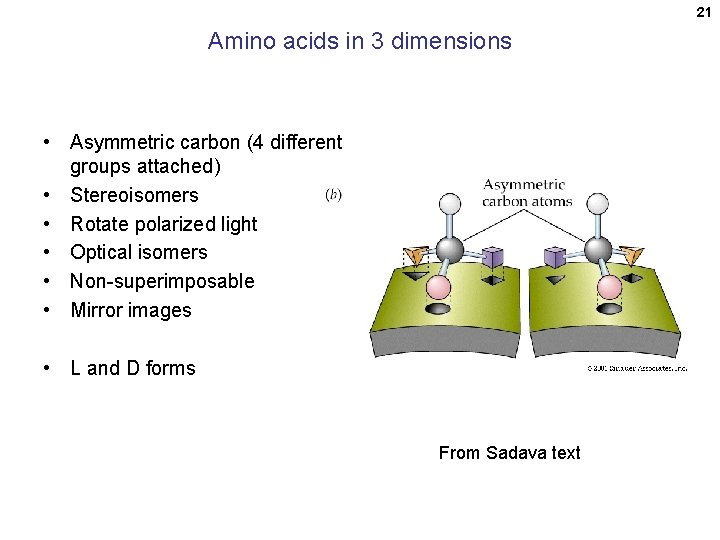
21 Amino acids in 3 dimensions • Asymmetric carbon (4 different groups attached) • Stereoisomers • Rotate polarized light • Optical isomers • Non-superimposable • Mirror images • L and D forms From Sadava text

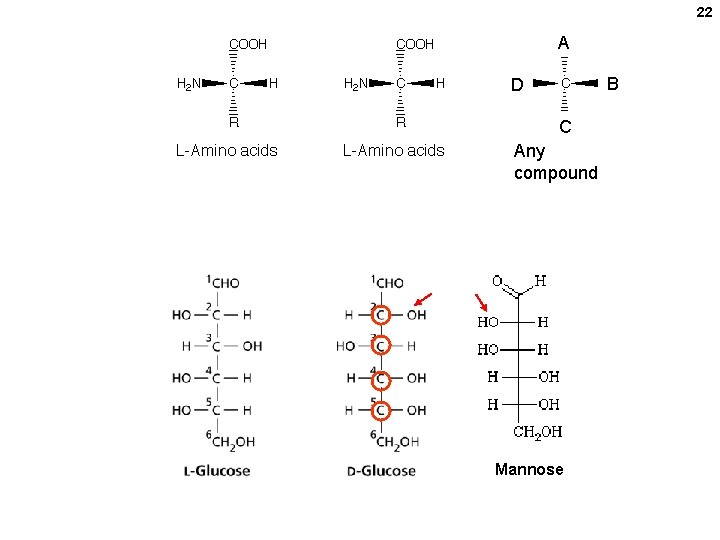
22 A B D C Any compound Mannose

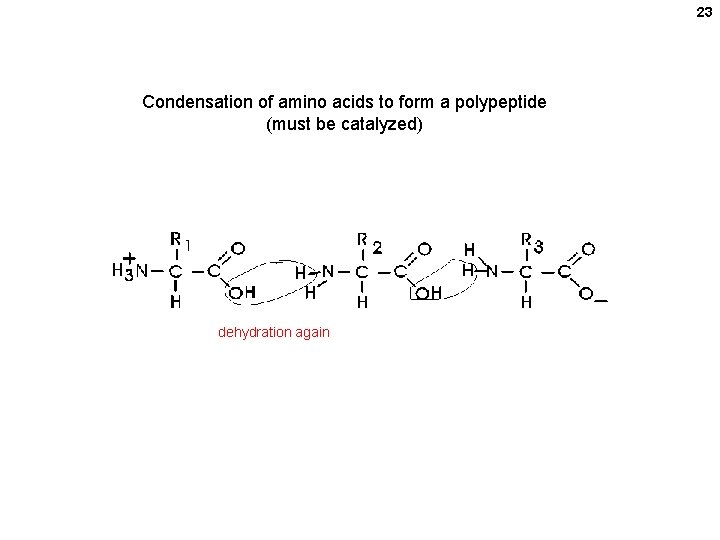
23 Condensation of amino acids to form a polypeptide (must be catalyzed) dehydration again

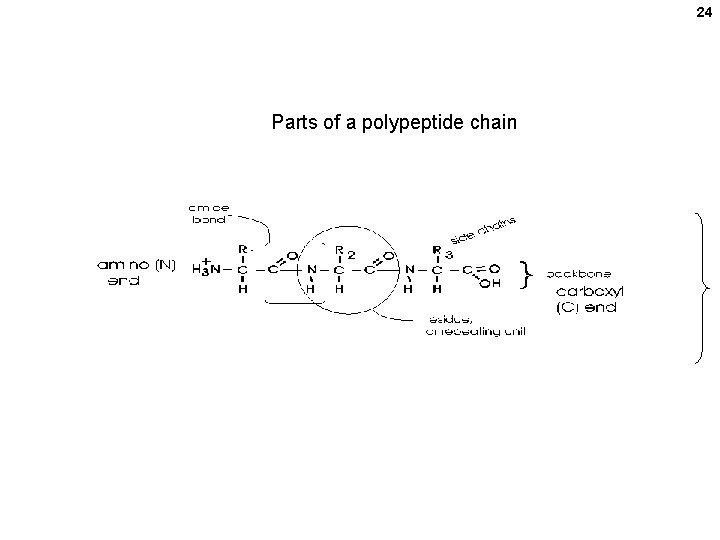
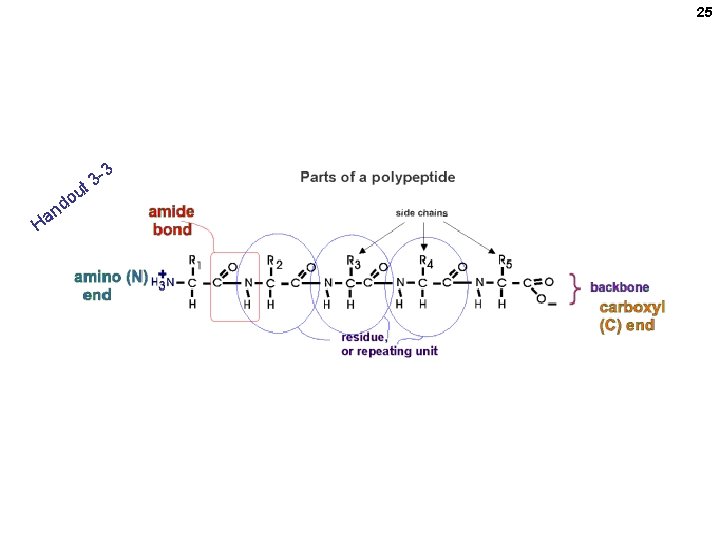
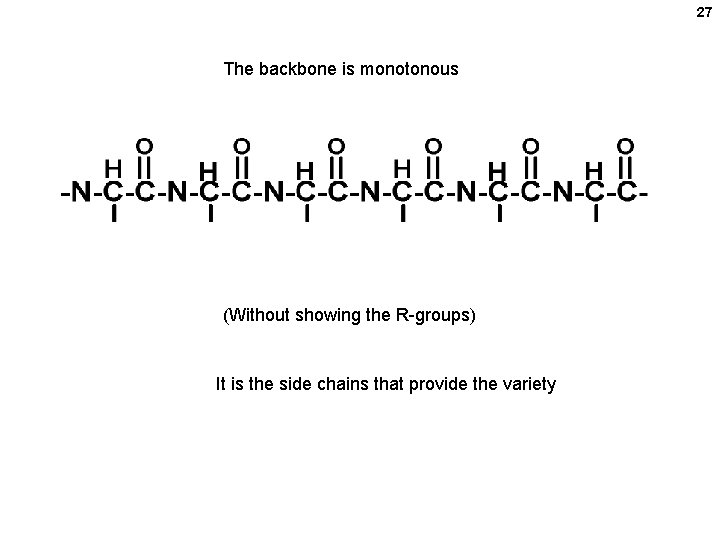
24 Parts of a polypeptide chain

25 3 t 3 u o nd Ha The backbone is monotonous It is the side chains that provide the variety

26 “Polypeptides” vs. “proteins” • Polypeptide = amino acids connected in a linear chain (polymer) • Protein = a polypeptide or several associated polypeptides (discussed later) • Often used synonymously • Peptide (as opposed to polypeptide) is smaller, even 2 AAs (dipeptide)

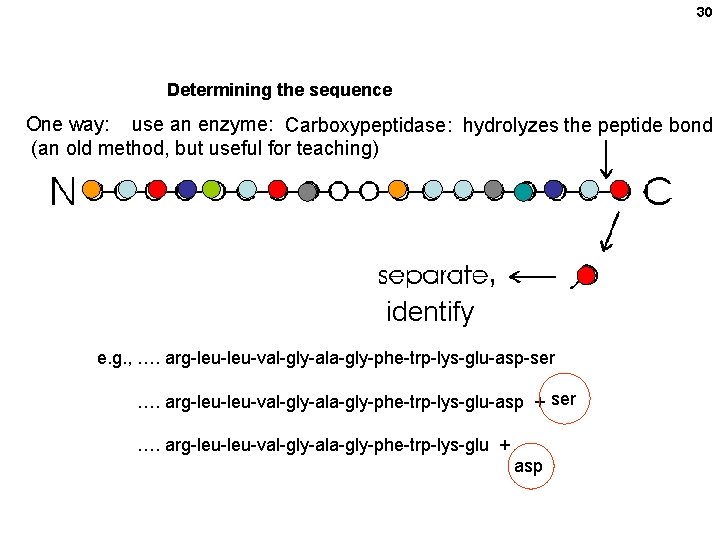
27 The backbone is monotonous (Without showing the R-groups) It is the side chains that provide the variety

28 Proteins do most of the jobs in the cell E. g. , egg albumin, hemoglobin, keratin, estrogen receptor, immunoglobulins (antibodies), enzymes (e. g. , beta-galactosidase) Each is a polymer or assemblage of polymers made up of amino acids Each particular protein polymer (polypeptide) has a unique sequence of amino acids. . and an English name. Each molecule of a particular protein has the same sequence of amino acids. E. g. , met-ala-leu-arg-glu-leu-val-. . How is this sequence determined?

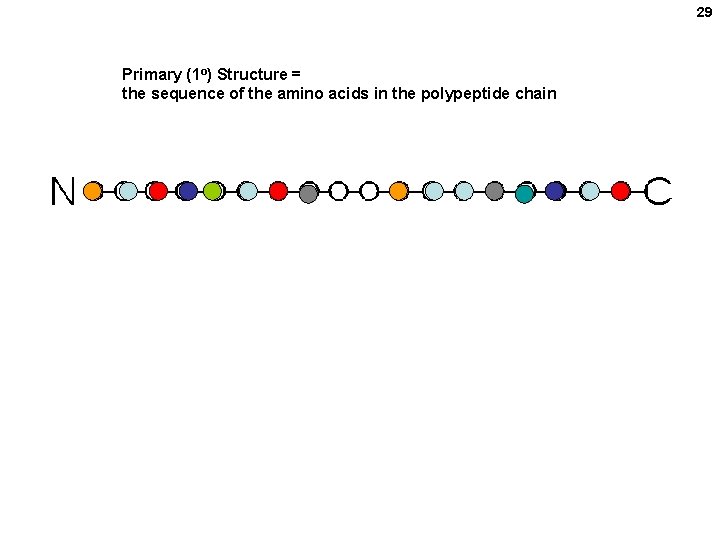
29 Primary (1 o) Structure = the sequence of the amino acids in the polypeptide chain

30 Determining the sequence One way: use an enzyme: Carboxypeptidase: hydrolyzes the peptide bond (an old method, but useful for teaching) , identify e. g. , …. arg-leu-val-gly-ala-gly-phe-trp-lys-glu-asp-ser …. arg-leu-val-gly-ala-gly-phe-trp-lys-glu-asp + ser …. arg-leu-val-gly-ala-gly-phe-trp-lys-glu + asp

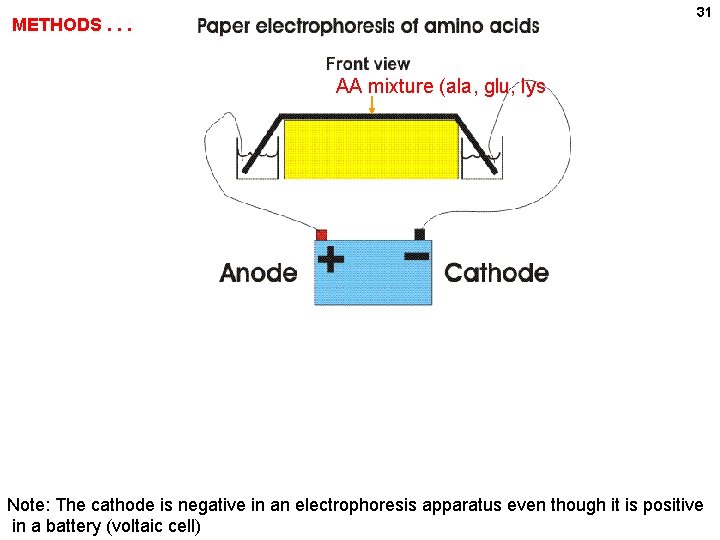
31 METHODS. . . AA mixture (ala, glu, lys Anode (-) (+) Cathode Note: The cathode is negative in an electrophoresis apparatus even though it is positive in a battery (voltaic cell)

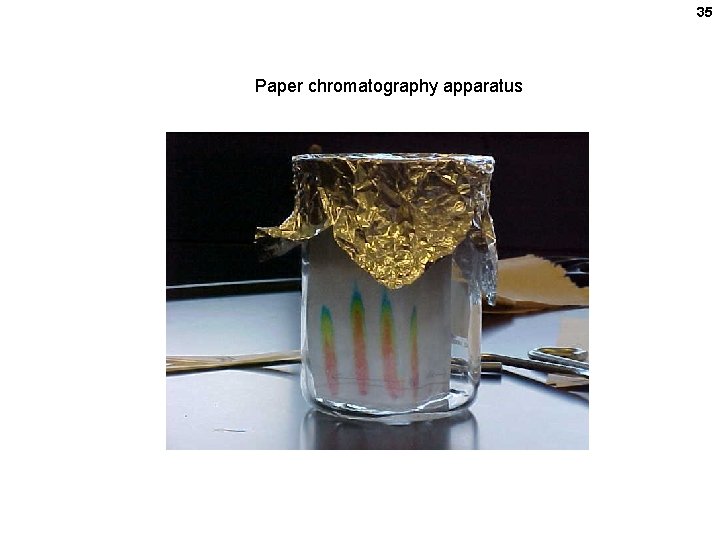
32 A paper electrophoresis apparatus

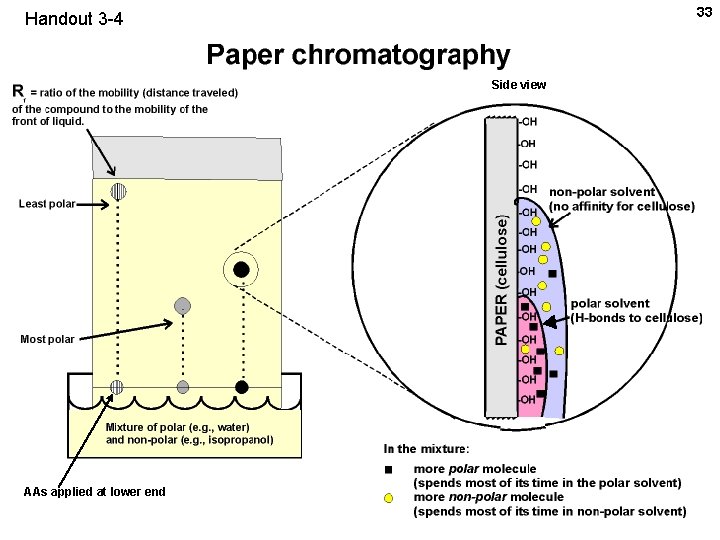
33 Handout 3 -4 Side view AAs applied at lower end

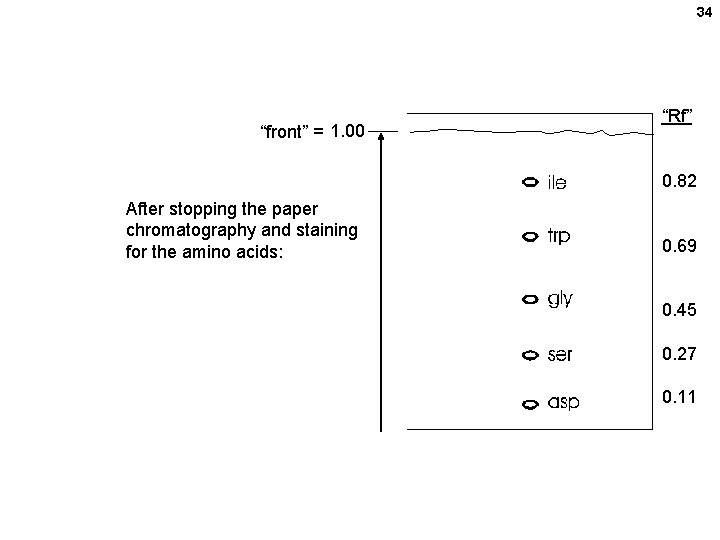
34 “front” = 1. 00 “Rf” 0. 82 After stopping the paper chromatography and staining for the amino acids: 0. 69 0. 45 0. 27 0. 11

35 Paper chromatography apparatus

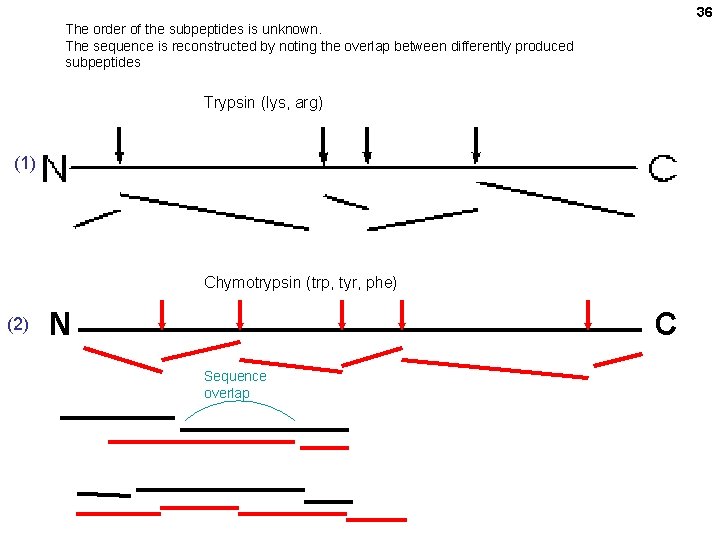
36 The order of the subpeptides is unknown. The sequence is reconstructed by noting the overlap between differently produced subpeptides Trypsin (lys, arg) (1) Chymotrypsin (trp, tyr, phe) (2) N C Sequence overlap

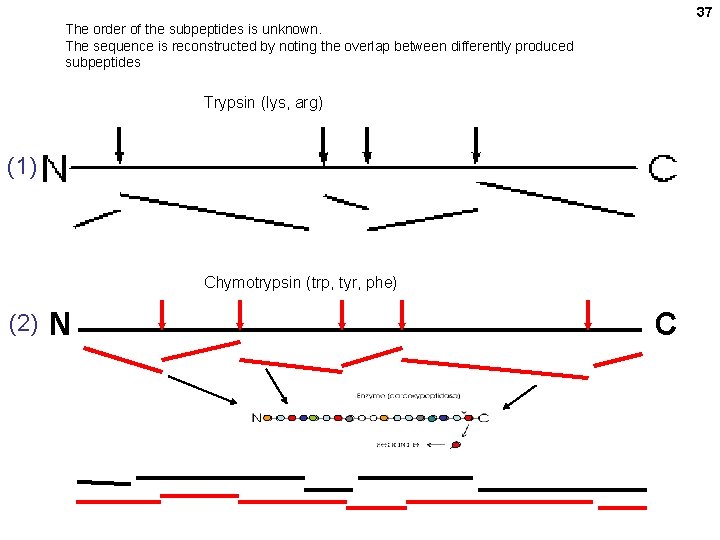
37 The order of the subpeptides is unknown. The sequence is reconstructed by noting the overlap between differently produced subpeptides Trypsin (lys, arg) (1) Chymotrypsin (trp, tyr, phe) (2) N C

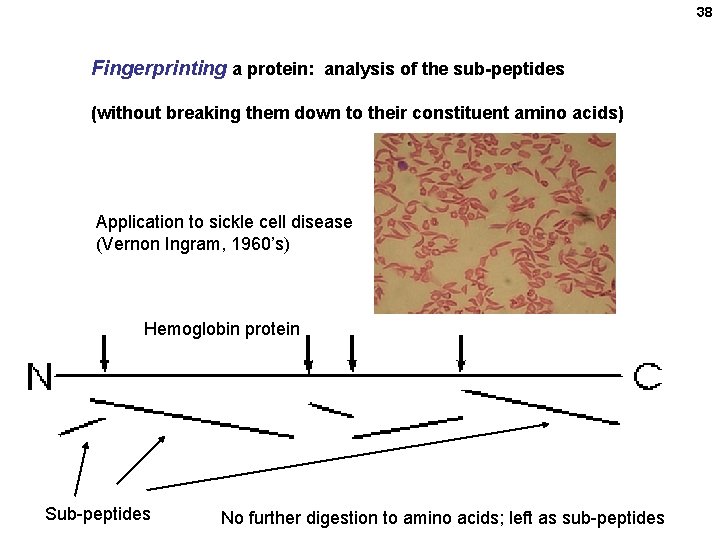
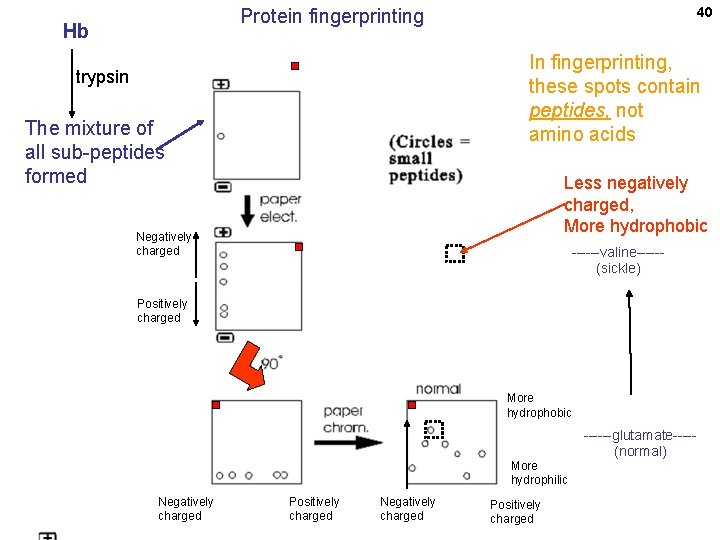
38 Fingerprinting a protein: analysis of the sub-peptides (without breaking them down to their constituent amino acids) Application to sickle cell disease (Vernon Ingram, 1960’s) Hemoglobin protein Sub-peptides No further digestion to amino acids; left as sub-peptides

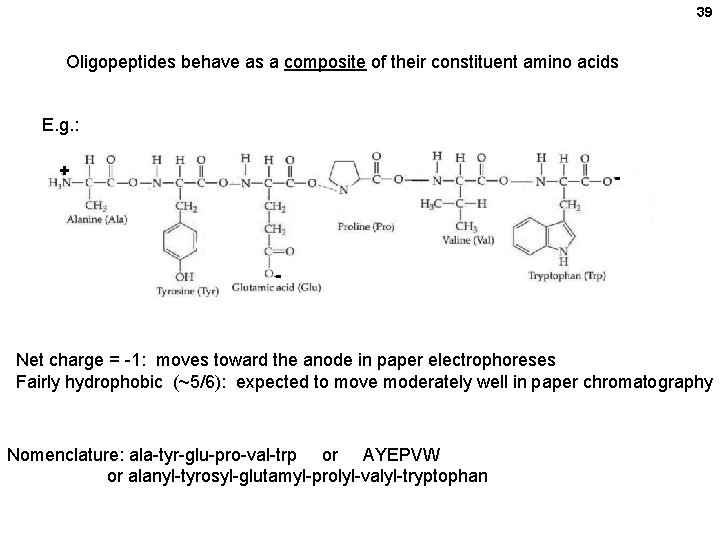
39 Oligopeptides behave as a composite of their constituent amino acids E. g. : + - - Net charge = -1: moves toward the anode in paper electrophoreses Fairly hydrophobic (~5/6): expected to move moderately well in paper chromatography Nomenclature: ala-tyr-glu-pro-val-trp or AYEPVW or alanyl-tyrosyl-glutamyl-prolyl-valyl-tryptophan

40 Protein fingerprinting Hb In fingerprinting, these spots contain peptides, not amino acids trypsin The mixture of all sub-peptides formed Less negatively charged, More hydrophobic Negatively charged ------valine-----(sickle) Positively charged More hydrophobic More hydrophilic Negatively charged Positively charged ------glutamate----(normal)

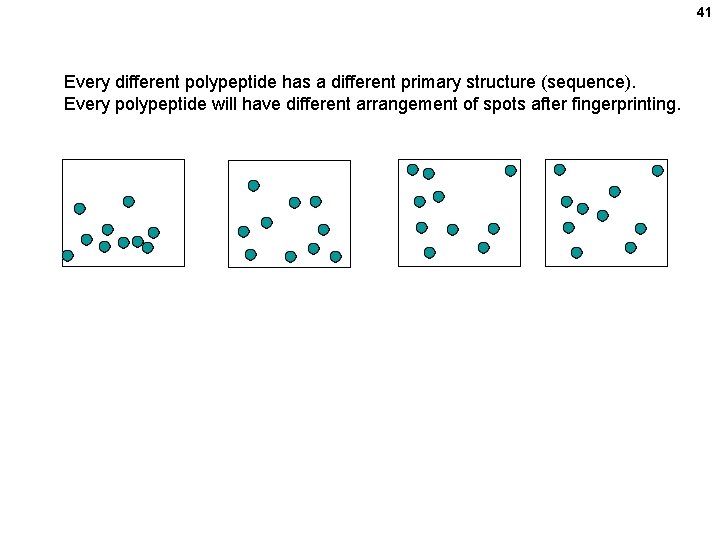
41 Every different polypeptide has a different primary structure (sequence). Every polypeptide will have different arrangement of spots after fingerprinting.

42 3 -dimensional structure of proteins One given purified polypeptide • Molecule #1: N-met-leu-ala-asp-val-lys-. . • Molecule #2: N-met-leu-ala-asp-val-lys-. . . • Molecule #3: N-met-leu-ala-asp-val-lys-. . . • Molecule #4: N-met-leu-ala-asp-val-lys-. . . etc. clothesline. . .

43 Information for proper exact folding (How does a polypeptide fold correctly? ) Predicting protein 3 -dimensional structure Determining protein 3 -dimensional structure Where is the information for choosing the correct folded structure? Is it being provided by another source (e. g, a scaffold) or does it reside in the primary structure itself?

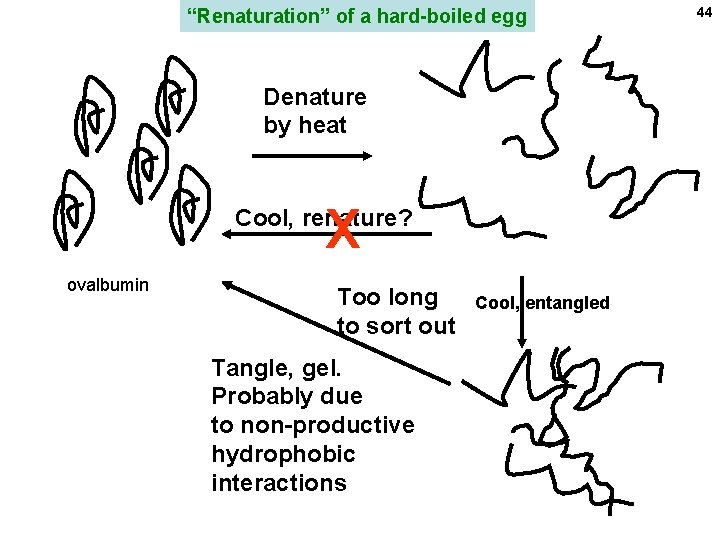
“Renaturation” of a hard-boiled egg Denature by heat X Cool, renature? ovalbumin Too long to sort out Tangle, gel. Probably due to non-productive hydrophobic interactions Cool, entangled 44

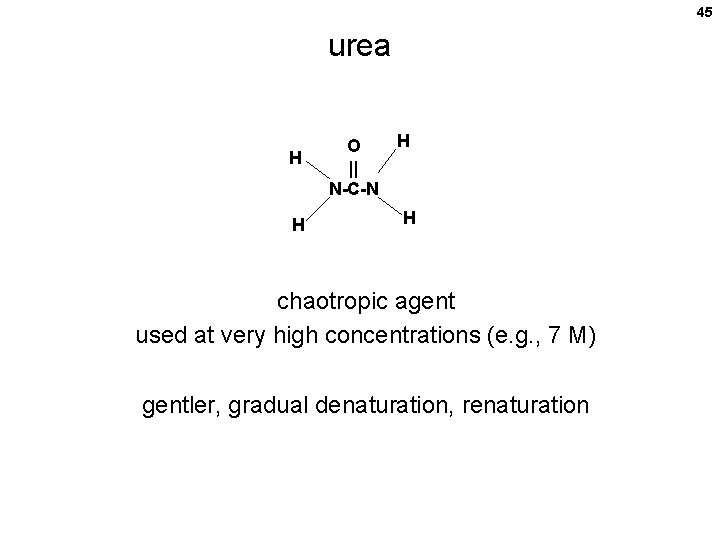
45 urea H H H O || N-C-N H chaotropic agent used at very high concentrations (e. g. , 7 M) gentler, gradual denaturation, renaturation

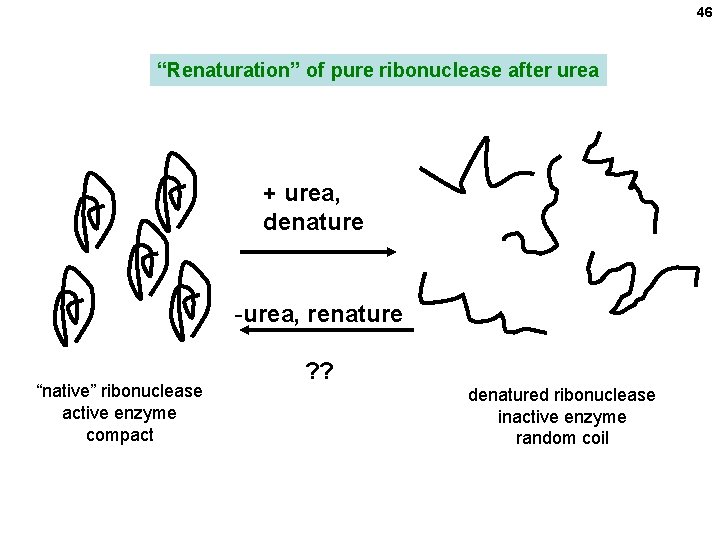
46 “Renaturation” of pure ribonuclease after urea + urea, denature -urea, renature “native” ribonuclease active enzyme compact ? ? denatured ribonuclease inactive enzyme random coil

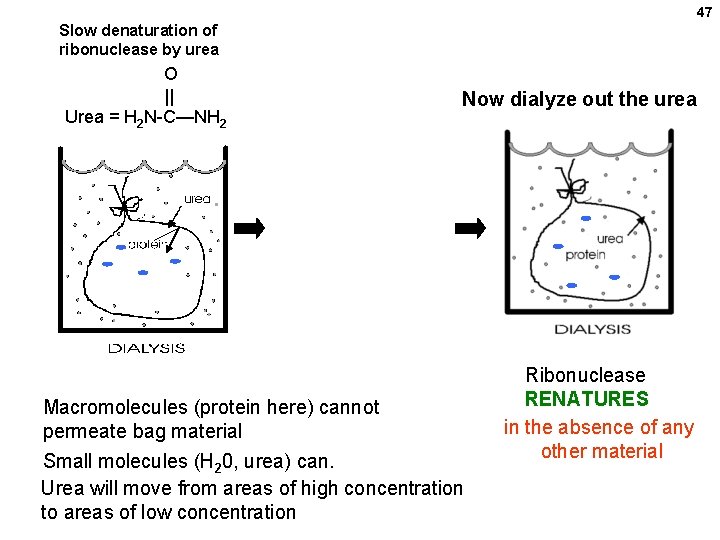
47 Slow denaturation of ribonuclease by urea O || Urea = H 2 N-C—NH 2 Ribonuclease in the bag is denatured Now dialyze out the urea Macromolecules (protein here) cannot permeate bag material Small molecules (H 20, urea) can. Urea will move from areas of high concentration to areas of low concentration Ribonuclease RENATURES in the absence of any other material

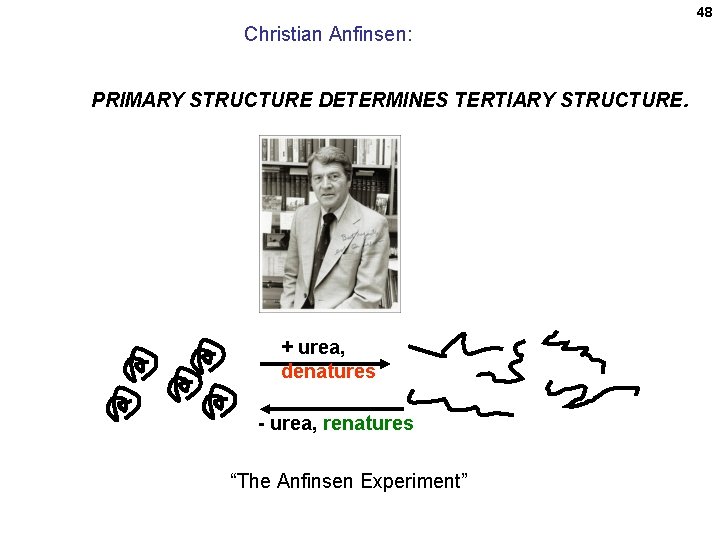
48 Christian Anfinsen: PRIMARY STRUCTURE DETERMINES TERTIARY STRUCTURE. + urea, denatures - urea, renatures “The Anfinsen Experiment”